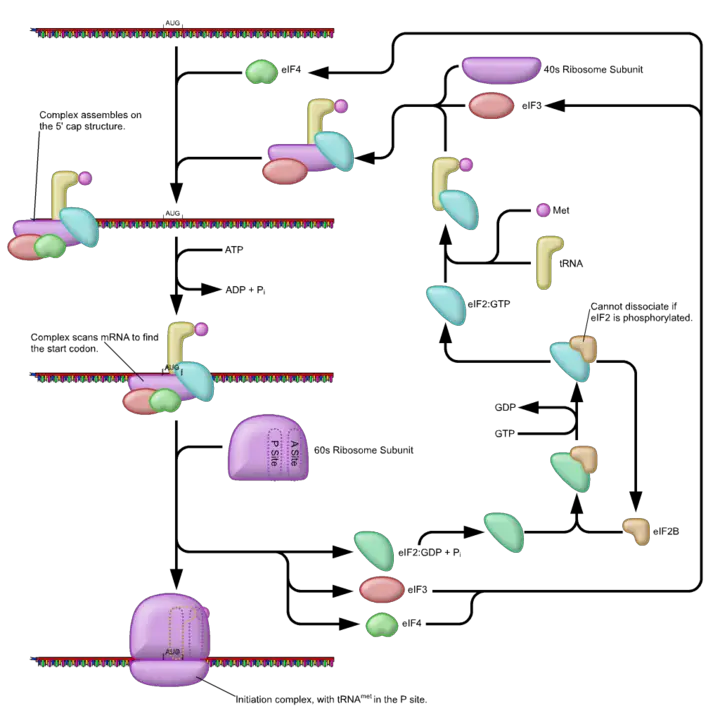Translation in Eukaryotes and Prokaryotes
Overview
Using a microscope in 1665, Robert Hooke discovered tiny units of cork tissue which reminded him of monastery cells (rooms) that monks inhabited. He, therefore, referred to these units as cells. However, what Hooke actually saw using his microscope were dead cell walls of the tissue. It was not until 1674 that Anton van Leeuwenhoek used a microscope to observe a live cell.
Today, it's widely believed that what Leeuwenhoek observed under the microscope was a bacterial cell. Along with other findings, these discoveries resulted in the formulation of the Cell Theory by Matthias Schleiden in 1839 which states that a cell is a basic unit of life (the theory also holds that new cells originate from existing cells and that all living things have one or more cells).
Today, cells are divided into two main categories namely, prokaryotic cells (Archaea and Bacteria) and eukaryotic cells (plants, animals, protists, etc). As the names suggest, the two types of cells are classified based on the manner in which their genetic material are arranged/organized within the cell. However, they also have a number of other differences that make it possible to distinguish between the two types of cells.
* The word nucleus is derived from the Latin word nucleus that means "kernel/core".
* Whereas "Eu" means true or good, "Pro" means no - Here, then, Eukaryotes may be described as cells that have a nucleus while prokaryotes are cells without a nucleus. However, it's worth noting that they all have genetic material.
Translation
In molecular biology and genetics, translation is the term used to describe the process through which a messenger Ribonucleic acid (mRNA) is decoded in order to synthesize polypeptides or amino acid chains. Here, mRNA carries genetic codes (information) that serve as the blueprint of these molecules (used to build proteins). In cells, this process occurs after transcription and involves three main stages.
These include:
- Initiation
- Elongation
- Termination
Apart from the differences in how genetic material are organized between eukaryotes and prokaryotes, differences can also be identified in translation between the two types of cells.
A Brief Description of Transcription in Prokaryotes and Eukaryotes
Given that mRNA, which serves as a template for proteins synthesis, is itself a product of transcription, it's important to get a general idea of this process in prokaryotes and eukaryotes.
* Transcription may be described as the process that connects DNA (or genetic information contained in DNA) to protein. Here, the information contained in DNA is ultimately used to produce proteins.
In eukaryotic cells, the transcription process takes place within the nucleus and the resulting mRNA transcript is transported to the cytoplasm where it's involved in translation. In prokaryotes, on the other hand, transcription takes place in the cytoplasm where the genetic material is located.
Here, it's worth noting that unlike eukaryotic cells, prokaryotes do not have a nucleus where the genetic material is bound by a membrane. As a result, genetic material of the cell is located in the cytoplasm.
In both eukaryotes and prokaryotes (bacteria), the first stage of transcription is known as the initiation stage and starts when associated proteins and enzymes (RNA polymerase) bind onto the promoter (a DNA sequence).
A good example of these sequences (at the promoter) is the TATA box in eukaryotes (this is an ideal site given that the As and Ts are bound by a few (2) hydrogen bonds and thus easier to pull the strands apart).
In eukaryotic cells, proteins known as basal transcription factors have to bind to the promoter site first in order to help the RNA polymerase attach to the site. This is different when compared to prokaryotes where the polymerase attaches to the promoter directly.
* During the initiation stage, binding of the polymerase to the promoter region results in the unwinding of the DNA before the second stage starts.
* In eukaryotes, the transcription factors (TFs) are important in that they identify and bind to the DNA sequence in the promoter region. Once they bind to the site, they form what is known as the initiation complex that attracts the polymerase to bind.
The next (second) stage of transcription is known as elongation and can simply be described as the elongation of the transcript. Here, the polymerase "reads" and "writes" the mRNA from the (-) antisense template strand of the DNA while the (+) sense strand protects it (the negative antisense template strand) from various interfering factors.
Given that the polymerase copies from the template strand, the mRNA being formed is complementary to this strand. However, this new strand contains a Uracil (U) nucleotide rather than a Thymine (T) present in the DNA strand.
* During elongation, the polymerase "moves" along the template strand in a 3' to 5' direction adding a nucleotide to the RNA in a manner that matches those of the DNA strand. This produces a transcript (RNA transcript) that is almost identical to the non-template.
The last phase of transcription is known as termination where transcribing continues until it's stopped which in turn allows the RNA transcript to be liberated. Here, the polymerase may be instructed to dissociate from the template by given termination signals depending on the cell.
In prokaryotes, protein-based signals such as rho protein control Rho-dependent termination which results in the polymerase dissociating from the template as the mRNA is liberated.
* Given that transcription occurs in the cytoplasm in prokaryotes, translation often starts while transcription continues or immediately after it ends. In eukaryotes, however, a nuclear membrane separates the ribosome (involved in the translation process) from the transcription process. For this reason, transcription has to be completed before the transcripts are released into the cytoplasm where translation takes place.
Characteristics of the mRNA of Prokaryotes and Eukaryotes
mRNA produced through the transcription process is also known as mRNA transcripts. Although they have a number of similar characteristics, they also have several differences. The prokaryotic mRNA transcript can be divided into a number of parts/sections that include: the non-coding region (located at the 5' end of the transcript), the Shine-Dalgarno sequence, a second non-coding region, the start codon, the coding region, stop codon and another non-coding region on the 3' end.
The eukaryotic mRNA, on the other hand, starts with a 5' cap and consists of a guanine nucleotide. This nucleotide is attached to a methyl group and bound to the neighboring nucleotide. The guanine nucleotide is attached to the non-coding region, similar to the one in prokaryotic mRNA. The next section is the start codon from which the coding region extends.
The coding region ends at the stop codon. This is followed by a non-coding region and lastly the poly-A-tail (made up of adenines and may consist of as many as 2200 nucleotides) at the 3' end. In eukaryotes, the 5' cap and the poly-A tail prevent the mRNA from being degraded.
Here, it's important to remember that in eukaryotes, the mRNA has to be released into the cytoplasm where translation takes place. Therefore, the two sections play an important role in maintaining the integrity of the mRNA. In prokaryotes, transcription and translation can occur at the same time and thus these sections are not necessary.
Unlike the eukaryote transcript, this mRNA does not have to be transported a long distance and thus does not encounter various enzymes that are likely to degrade it. As a result, the mRNA in prokaryotes does not require additional protection to prevent damage.
As mentioned, translation is the process through which the building blocks of proteins (polypeptides/amino acid chains) are built using the information contained in the mRNA. It's an important process given that it produces proteins that are required for various cell functions.
In order to understand the process it is important to know some of the components and terminologies used in translation.
Apart from mRNA (messenger RNA), they include:
· Polypeptides - Chains of amino acids and are the molecules that make up proteins.
· Nucleotides - Structural components of DNA and RNA. They are themselves made up of nucleoside and phosphate and include adenine, thymine, cytosine, and guanine (as well as Uracil).
· Codons - A group consisting of three nucleotides - For instance, AUG is a good example of a codon - While codons serve as the building blocks of amino acids, others stop the process once the polypeptide is complete.
· tRNA (transfer RNA) - Act as the bridge between mRNA codons and amino acids.
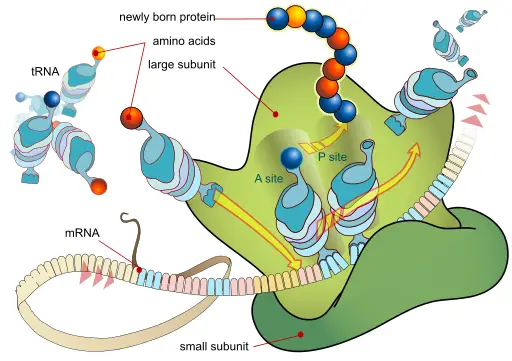
· Ribosome - Ribosome consist of rRNA, and protein and are the structures in which polypeptides are manufactured.
Translation in Prokaryotes
Given that the genetic material (DNA) of prokaryotes is not contained in a membrane-bound nucleus, transcription takes place in the cytoplasm. This, then, allows translation to begin in this environment as soon as mRNA emerges from the polymerase (RNA polymerase/RNAP).
In cases where is there is enough room (on the mRNA) to accommodate the ribosome, translation can start even before the transcription process is completed.
As a result, a scenario in which a strand of DNA is being transcribed by multiple polymerases with multiple ribosomes translating this information (of the RNA) is not unusual in prokaryotes particularly when it comes to highly expressed genes.
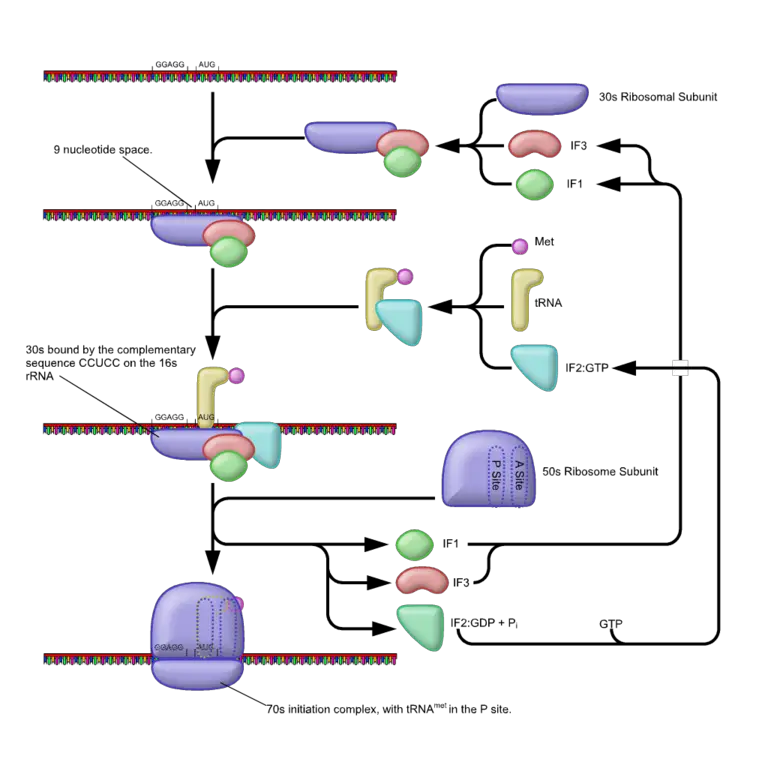
As is the case with transcription, there are three phases of translation which include initiation, elongation, and termination. The initiation phase is characterized by formation of the initiation complex and starts with the small subunit of the ribosome (30S) binding to the mRNA.
* Ribosome consists of two subunits (rRNA subunits) with one of the subunits being smaller than the other. In prokaryotes, the smaller subunit is designated 30S while the larger one is 50S - the total of these is 70S (S stands for Svedberg units.)
Initiation
For the initiation phase to take place, the smaller ribosomal subunit first has to be dissociated from the larger (50S) ribosomal subunit. Once it's dissociated, initiation factors (IF-1 and IF-2) bind at given sites on the 30S subunits where they serve different functions.
At the A site (of the ribosome subunit), IF-1 serves to prevent a new molecule of aminoacyl-tRNA from entering at this stage of translation. In addition, it promotes the assembly and stabilization of the complex.
As well, the IF-3 initiation factor promotes binding of the subunit to mRNA. The third initiation factor (IF-2 GTP) introduces the initiator aminoacyl-tRNA and binds the P site of the subunit. In doing so, it allows the anticodon of the tRNA to attach to the start codon (AUG) of the mRNA.
Following hydrolysis of the GTP (as well as the release of the other initiation factors) the larger ribosome subunit (50S) binds to the smaller subunit (30S) which produces a fully functional ribosome. Following the formation of a fully functional ribosome, the A site can again accept another aminoacyl-tRNA molecule.
By the end of the initiation phase, the initiation complex that is formed consists of both ribosomal subunits (the large and smaller subunits), the mRNA, as well as the tRNA that also carries fMet (N-formyl-methionine).
* IF-1 and IF-3 also help dissociate the smaller ribosome subunit (30S) from the larger subunit (50S).
* The Shine-Dalgarno sequence is located several bases upstream of the start codon (in the mRNA). This site is important in that it signals the protein synthesis process by properly aligning the ribosome subunit to the start codon.
* tRNA, which is one of the initiators carries N-formyl-methionine (fMet) which is inserted into the N terminus of the polypeptide chains produced by such prokaryotes as E.coli.
Elongation
The second phase of translation is known as elongation and is characterized by the elongation of the polypeptide chain. Here, the ribosome has a catalytic function as a peptido-transferase.
The entire process can be divided into three main steps of the elongation that include: aminoacyl-tRNA binding, formation of the peptide bond, as well as translocation. During the first step of this cycle (aminoacyl-tRNA binding), an aminoacyl-tRNA that corresponds to the second codon binds to the A site (aminoacyl site) through the codon-anticodon interaction.
Here, it's worth noting that methionine that came with the IF-2 along with the initiator tRNA during the initiation phase is the first amino acid. Binding of the aminoacyl-tRNA is promoted by GTP and the elongation factor (ET-Tu). The three join to form a complex (aminoacyl-tRNA/EF-Tu/GTP complex) which results in the hydrolysis of GTP. In turn, the elongation factor (EF-Tu bound t GDP) is released.
The released EF-Tu molecule can then promote the binding of another tRNA to the ribosome once it has regenerated. This occurs when EF-Ts (also an elongation factor) binds and replaces the GDP on the EF-Tu. The EF-Ts are then replaced by GTP resulting in the formation of a newly regenerated EF-Tu-GTP.
In the second step, forming the peptide bond, the carboxyl end of the amino acid on the tRNA at the Peptidyl site (P) is dissociated and binds to the amino group of the amino acid that is joined to the tRNA at the A site through a peptide bond. This step of the cycle is catalyzed by the peptidyl transferase.
The third step of the cycle (translocation) is characterized by binding of the elongation complex and GTP to the ribosome. Here, hydrolysis of the GTP results in the production of GDP and a phosphate while the release of the elongation factor (EF-G) frees it to bind GTP in preparation of another elongation cycle.
With the deacylated tRNA moving from the P site to the E site and the dipeptidyl tRNA from the A to the P site, the site remains empty and thus free to accept another aminoacyltRNA. An amino acid is continually added to the C terminal end of the polypeptide as it grows in length for each of the codons as the peptidyl-tRNA moves to and from the P and A sites.
Termination
* During elongation, the tRNA continually moves from the P to the A site (forwards) as it brings the next amino acid to be added onto the previous chain (chain that started with a methionine). This process continues until a stop codon in the mRNA enters the A site thus stopping the cycle from continuing. There are three types of stop codon which include; UAA, UAG, and UGA.
The last phase of the translation process is known as termination and is the point at which the process ends. Having entered the A site, the stop codon prevents tRNA from binding.
One of the release factors (RF-1 or RF-2 along with a RF-3) binds to the codons causing enzyme (peptidyl transferase) responsible for peptide bonds to release a water molecule onto the last amino acid on the chain which causes the peptide and tRNA attached to the P site to be hydrolyzed. As a result, the newly formed chain is separated from tRNA and leaves the ribosome.
* Whereas RF-1 identifies UAA and UAG, RF-2 identifies UAA and UGA while RF-3 promotes the interaction of either of the other two release factors with the ribosome.
* Release factors bind to the stop codon given that no tRNA have anticodon for the stop codon in prokaryotes.
Some of the other events that take place during the termination phase include:
· mRNA is released
· tRNA is released from the ribosome when ribosome releasing factor binds onto the A site
· The ribosome dissociates into the large and small subunits when EF-G binds to the RRF (ribosome releasing factor)
Translation in Eukaryotes
As is the case in prokaryotes, translation is the process through which a sequence of mRNA is translated into polypeptides during protein synthesis.
As mentioned, transcription and translation processes occur in the cytoplasm in prokaryotes (and can even occur at the same time). However, in eukaryotes, the nucleus membrane separates the ribosome located in the cytoplasm from the transcription process that takes place in the nucleus. For this reason, translation starts when transcription ends and the mRNA is transported to the cytoplasm.
* To reach the cytoplasm, mRNA passes through the nuclear pores on the nuclear membrane.
* In eukaryotes, translation also occurs in ribosome located on the Endoplasmic Reticulum (ER).
In eukaryotic organisms, translation also occurs in three phases that include initiation, elongation, and termination. While this is similar to the process in prokaryotes, there are several differences particularly with regards to the components involved.
Initiation
During the initiation phase, the smaller ribosomal subunit forms a complex with three initiation factors. Here, however, the smaller ribosomal subunit is 40S compared to the much smaller 30S in prokaryotes. Binding of these initiation factors (IF-1, IF-A, and IF-3) to the ribosomal subunit produce the preinitiation complex that in turn joins the IF-5 (initiation factor 5) and tRNA.
Ultimately, this complex binds the mRNA to form the initiation complex. As is the case in prokaryotes, the small ribosomal subunit moves along the untranslated region of the mRNA as it scans for the start codon (in most cases, the first AUG serves as the start codon in eukaryotes).
* In eukaryotes, the mRNA sequence located at the start codon is known as the Kozak sequence (ACCAUGG). While this sequence serves a similar function to the Shine-Dalgarno sequence, the two are different in that the Kozak sequence actually contains the start sequence.
Once the start codon is recognized, the larger ribosome (60S) subunit is recruited to the complex which results in the formation of a fully functional ribosome (this is an energy-dependent process that involves GTP hydrolysis and ultimately produces an 80S ribosome). Once a fully functional ribosome is formed, the initiation factors are released.
* At the end of the initiation factor, the initiator tRNAmet is located at the P site while the A site remains vacant.
Elongation
This is the second phase of translation and involves the synthesis of polypeptide. While the elongation process in eukaryotes is similar to that of eukaryotes, EF-Tu is replaced by EF-1α. Here, the elongation factor proteins (EF) has three main functions.
The first function of these proteins (elongation factor proteins) is to recruit the charged tRNAs to the A site. In addition, they play an important role in forming a peptide bond between the amino acids as well as translocation of the ribosome along the mRNA.
Progress of the process involves the translocation event. In each of these events, the charged tRNAs enter the A site before shifting onto the P site. At the end of the each event, the tRNA enters the E site so that it can be removed.
As the ribosome moves along the mRNA, elongation factors promote the peptide bonds between amino acids located at on tRNA (at the A site) and the carboxyl group of the amino group that is located on the tRNA of the P site.
Here, peptidyl transferase (ribozyme located in the larger 50S ribosomal subunit) serves to catalyze the reaction. The amino acid associated with the tRNA on the P site is then linked to the growing polypeptide chain which allows the chain to continue growing in length. This process allows the ribosome to continue moving along the mRNA as the polypeptide chain continues growing before it stops at the termination phase.
Termination
This is the last phase of the translation process. It occurs when the ribosome arrives at the nonsense codon of the mRNA where the tRNA has no complementary anticodon. Once the nonsense codon is identified by release factors, the amino acid at the P site is detached from the tRNA which frees the polypeptide.
On the other hand, the ribosome is not only dissociated from the mRNA, but also into the two subunits (small and large ribosomal subunits) which allows them to enter the initiation phase in another translation process.
Return from Translation in Eukaryotes and Prokaryotes to MicroscopeMaster home
References
Birge E.A. (2000) Transcription and Translation: Processes and Basic Regulation. In: Bacterial and Bacteriophage Genetics.
Eric Wong. (2009). Cells: Molecules and Mechanisms: Translation :From RNA to Protein.
Pelin Pelit Arayici, Tayfun Acar, and Mesut Karahan. (2014). Transcription and translation.
Julie A Theriot. (2013). Why are bacteria different from eukaryotes?
Suzanne Clancy & William Brown. (2008). Translation: DNA to mRNA to Protein.
Links
https://openstax.org/books/biology-ap-courses/pages/15-2-prokaryotic-transcription
https://openoregon.pressbooks.pub/mhccmajorsbio/chapter/prokaryotic-transcription/
https://courses.lumenlearning.com/boundless-biology/chapter/prokaryotic-transcription/
Find out how to advertise on MicroscopeMaster!