Histones and their Functions
Overview: What are Histones?
With an average diameter of about 10nm, histones are chromatin proteins on which DNA is wrapped and compacted within eukaryotic cells (and Archaea species).
Histones are divided into two main categories, linker histones (H1 and H5) and core histones (H2A, H2B, H3, and H4).
While histones play an important role in DNA packaging, where DNA is wrapped around histones, they are also involved in several other functions that include:
- Shaping the chromosomes
- Regulate gene activities
- Chromatic regulation
- Protect DNA
* A chromatin comprises DNA and proteins. These proteins include histones and non-histones. Unlike histones, non-histone proteins are involved in such activities as histone gene transcription and expression as well as scaffolding.
* The nucleosome is the first level of chromatin organization. It consists of a segment of DNA tightly wrapped around the core histones (H2A, H2B, H3, and H4).
* When the H1 protein binds to a nucleosome, the resulting structure is known as a chromatosome.
See more about DNA and Chromosomes.
Characteristics of Histones
Where Are Histones Located?
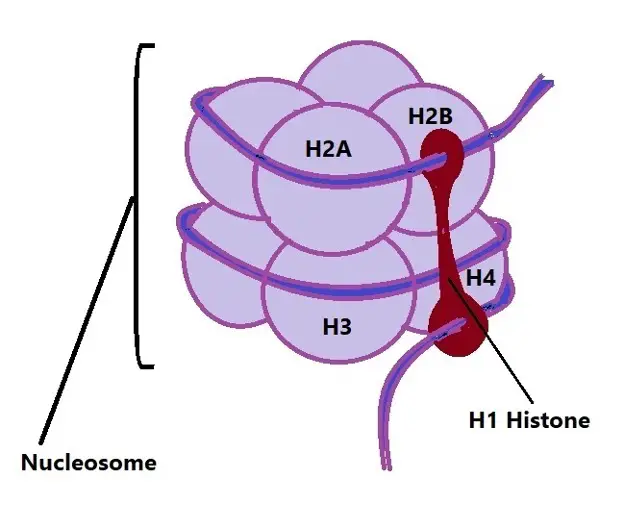
As mentioned, histones can be found within the nucleus of eukaryotic and archaea cells. Here, the linker histone (H1) is positioned near the dyad axis of the nucleosome. The dyad axis is the entering and exiting point at which DNA strands cross.
Core histones (consisting of a pair of H2A, H2B, H3, and H4), on the other hand, form the histone octamer and are located at the core of the nucleosome.
Diagrammatic representation of H1 attached to a nucleosome:
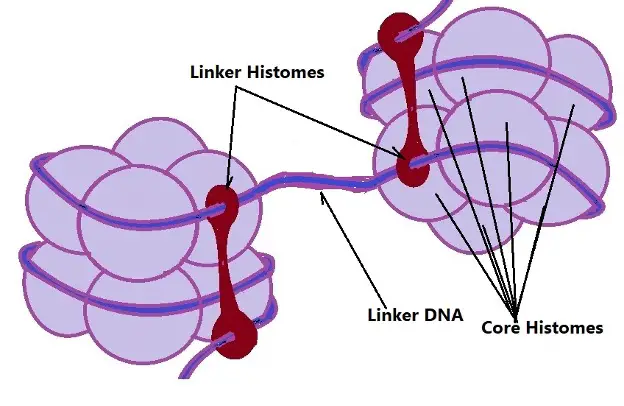
As shown in the diagram, a nucleosome consists of 8 core histones (H2A, H2B, H3, and H4 in a pair) and a segment of DNA tightly wrapped around them. While the H1 histone is located outside the core, it plays an important role in binding the DNA that enters and leaves the nucleosome. Here, they are also associated with the linker DNA (the segment of DNA between individual nucleosomes).
Some of the factors involved in nucleosome assembly are histone chaperones. At the beginning of the assembling process, some of these factors bind to certain histones in order to prevent nonspecific DNA association. The chaperones then guide the rest of the assembling process/path which hinges on a thermodynamic balance between histones binding to specific chaperone factors, association with DNA, as well as sub-nucleosomal structures. At the end of the assembling process, the resulting nucleosome is characterized by a disc-like complex.
Here, core histones H3 and H2A have been shown to interact with similar histones. Cationic tails of the structure are recognition motifs for chromatin binding proteins like HMGN-1 and the target of posttranscriptional modifications.
Though the histone tails act as regions of interaction for chromatic factors of given protein factors, fold regions of the histones have also been to serve the same purpose.
Structure and Organization of Histones
In eukaryotic organisms, linker histones (H1 Histone) generally consist of three main parts, the N-terminal tail, a central globular domain (with a winged-helix motif), and a lengthy C-terminal tail (about 100 aa in length).
Like the linker histone, core histones also consist of a central globular domain as well as an N-terminal tail (Only H2A has a long C-terminal tail). However, they vary in the number of amino acids present and the weight.
- H2A - 13.990- 14.0 kDa in weight. A H2A histone may consist of about 130 amino acids
- H2B - 13.8 kDa in weight with 126 amino acids
- H3 - 15 kDa
- H4 - 11kDa
* These figures represent the weight and number of amino acids in human beings.
* Core proteins/histones have a high content of lysine and arginine amino acids.
Diagrammatic Representation of Linker DNA:
Functions of Histones:
DNA Packaging
If stretched end-to-end, it's estimated that the DNA within a single human cell would be about 2 meters in length. However, within the cell, it has to be tightly packaged/compacted to fit within the nucleus which is about 2 micrometers in diameter.
* If the DNA in all the cells (of a single individual) were stretched and put together end-to-end, it's estimated that it would be twice the diameter of the solar system (or go to and from the sun over 300 times).
* A single cell contains 5 billion base pairs of DNA.
To be tightly compacted/packaged within the nucleus, the DNA strand is first wound around the histones.
While the addition of proteins (histones) might sound counterproductive, histones also serve to provide the energy required to effectively fold the DNA strand.
Moreover, studies have shown that chromatin (DNA-protein complex) can be easily packaged into a smaller volume as compared to DNA strands alone (without the protein).
As a result of the phosphate-sugar backbone, the DNA is generally negatively charged. As well, the high content of lysine and arginine amino acids in the core proteins confers an overall positive charge to the octameric core. As a result, these proteins bind the DNA tightly by means of electrostatic interactions.
Though the H1 histone is located outside the core proteins, it also binds and locks the DNA on the surface of the octomer. Here, it's worth noting that DNA is wound 1.65 times around the histone octamer. By linking to the DNA, the linker histone locks the strand in place along the length of the core and contributes to its packaging.
* In human beings, 147 bp (base pairs) of DNA wrap around the core histones 1.67 turns to form the nucleosome.
* The H1 histone wraps an additional 20 base pairs to complete two full turns around the core histones. At this point, the complex is known as a chromatosome with about 167 base pairs.
* The linker DNA contains about 20 base pairs.
Wounding DNA around histones shortens the strand sevenfold. However, even at this point, the chromatin is still too large to fit in the nucleus. For this reason, it has to be further coiled into what is known as 30-nanometer chromatin fiber.
While there is still a debate as to how the nucleosomes interact to form the fiber, studies have revealed that the H1 histones play a critical role in the process. In particular, the histone has been shown to help in contributing to the proper formation of the structure and stabilize the complex.
In addition to enhancing the stability of the fiber, the H1 histone is also involved in the formation of the helical twist of the fiber. Here, the interaction between the linker histones (H1 histone) in addition to interactions between nucleosomal interfaces in the structure ultimately results in the helical twist.
Transcriptional Activation and Inactivation
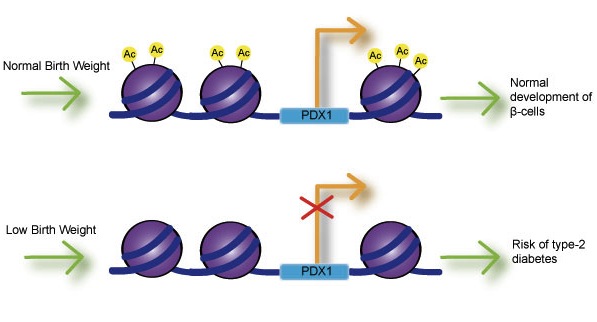
Histones are also involved in the activation and inactivation of transcriptional processes. As mentioned, histones have tails/extensions known as C-terminal and N-terminal tails. Here, post-translational modifications on the structure largely occur in the N-terminal tail and may involve methylation, Acetylation/phosphorylation, ubiquitylation, and SUMOlylation.
Histone Acetylation
The acetylation process results in the formation of the euchromatin which in turn allows for DNA transcription (gene expression). In this case, the introduction of the acetyl group (acetylation) reduces the positive charge of the histones so as to reduce the attraction between the positively charged histone and negatively charged DNA.
Specifically, the acetyl group targets and binds the lysine (responsible for the positive charge) and neutralizes it. By reducing the positive charge of lysine, the tight interaction between DNA and histones is reduced allowing the DNA to open up. Opening up the strand exposes the DNA to transcriptional machinery for transcription to occur.
* The enzyme histone acetyltransferases is responsible for linking the acetyl group to lysine to neutralize the positive charge. As well, the enzyme histone deacetylase is responsible for removing the acetyl group.
* The acetyl group may also serve as a docking site for proteins like bromodomains which are also involved in opening up the chromatin.
Histone Methylation
Unlike the acetylation process, histone methylation does not necessarily alter histone charge. Rather, methylation promotes transcriptional activity or causes inactivity. Here, however, it's worth noting that the methyl group also attaches to the lysine.
Lysine 4, located on histone 3 is involved in gene activity. Therefore, the methylation of this lysine on the histone activates the transcription process.
On the other hand, lysine 9 and lysine 27 on histone 3 (H3) are associated with inactivity. For this reason, methylation of these lysines results in inactivity.
Inactivity maintains or restores the heterochromatin structure where the chromatin does not open up to expose DNA to transcriptional factors. However, the type of heterochromatin is largely dependent on the type of lysine.
For instance, whereas lysine 9 is associated with constitutive heterochromatin, lysine 27 is associated with facultative heterochromatin.
* Methylation of lysine is carried out by specific lysine methyltransferases while lysine demethylases remove the methyl group.
* Lysine might be bound by a single methyl (Monomethyl lysine), two methyl groups (Dimethyl lysine), or three methyl groups (Trimethyl lysine). Each type of methylation has a given impact on specific genes.
* For example, when lysine 4 in histone 3 is dimethylated at the chromodomain, then a specific gene is ready for transcription. However, once the same lysine is trimethylated, the gene is activated because gene transcription is activated (the lysine has to be acetylated).
* Lysine methylation creates binding sites for co-activators.
Histone Phosphorylation
Generally, phosphorylation occurs on the amino acids tyrosine, serine, threonine, and histidine of the N-terminal tails. On histone 3 (H3), kinase enzymes are involved in the phosphorylation of serine and threonine. This results in the demethylation of the histone at position 9 which in can turn activates transcription.
For instance, the phosphorylation of serine 10 of histone 3 has been shown to cause demethylation of lysine 9 of the same histone. This allows for the acetylation of the lysine and consequently activates the transcription of specific genes.
* Whereas a kinase adds a phosphate group, the group is removed by a phosphatase enzyme.
* Serine, tyrosine, and threonine have a hydroxyl group that allows the phosphate to bind.
* During the normal cell cycle, serine 10 of histone 3 starts to be phosphorylated at the prophase stage. Metaphase is characterized by increased phosphorylation of the serine (peak phosphorylation) which declines in the next two stages (anaphase and telophase).
Given that the serine is only phosphorylated during some stages of the cell cycle, it has been postulated that the process serves to prepare the cell for segregation during the M-phase. Here, the phosphorylation of the serine helps prevent the aggregation of H3 tails thus allowing for increased compression of the chromosomes.
DNA Repair/DNA Damage Repair and Chromatin Remodeling
Histones (specifically H2AX) also play an important role in DNA damage repair and chromatin remodeling. This involves the phosphorylation of serine 139 of the variant histone (In mammals).
Unlike the modification of serine 10 of histone 3, this type of posttranslational modification occurs in all the phases (G1, S, G2, M) of the cell cycle to effectively respond to different forms of DNA damage.
Following DNA damage, studies have shown the serine (serine 129 in yeast and serine 139 in mammalian cells) to undergo phosphorylation under the action of protein kinases yTe11and yMecl.
In mammalian cells, the modified histone (known as yH2AX) spreads over several megabases of the break creating a signaling platform that recruits or helps retain DNA damage repair and signaling factors (e.g. NuA4 acetyltransferase complex). The recruitment of NuA4 at the site activates the hyperacetylation of H4 which in turn promotes the relation of local chromatin.
The modified histone helps in the recruitment of complexes like ATP-dependent remodelers INO80 and SWR1 that facilitate the repair process. On the other hand, factors like Rad9 act by delaying the progression of the cell through the G1 through to the S phase so as to allow enough time for efficient DNA repair.
* Once the repair process is complete, the modified histone (γH2AX) has to be removed from the affected site in order to prevent the retention of proteins involved in the repair process at this site. This not only stops the repair process but also allows the cell to process through the rest of the cell cycle phases.
* Here, some of the processes involved in the removal of γH2AX include replacing γH2AX with Htz1 histone variant as well as the dephosphorylation of the variant.
Return from "Histones and their Functions" to MicroscopeMaster home
References
Christopher L. Woodcock, Arthur I. Skoultchi & Yuhong Fan. (2006). Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length.
Leonardo Mariño-Ramírez, Maricel G Kann, Benjamin A Shoemaker, and David Landsman. (2005). Histone structure and nucleosome stability.
Marissa Vignali & Jerry L. Workman. (1999). Location and function of linker histones.
Links
https://www.sciencedirect.com/science/article/abs/pii/095506749190070F
https://medlineplus.gov/genetics/understanding/basics/chromosome/
https://www.researchgate.net/figure/Histone-location-and-function_fig4_264798727
Find out how to advertise on MicroscopeMaster!