Endonucleases Vs Exonucleases
Activities, Differences and Functions
Endonucleases and exonucleases are nucleases. As such, they are involved in the cleavage of phosphodiester bonds (between deoxyribose and phosphate residue) in nucleic acids. Generally, exonucleases serve to cleave the bonds located near the ends of the nucleic acid strands while endonucleases cleave bonds located within the strand. Some enzymes serve the same functions.
Despite their differences, nucleases have a number of important functions that include:
· Repair of nucleic acids
· Replication of nucleic acids
· Excision repair of nucleotides
· Repair of any mismatches
Endonuclease: Activities and Functions
As mentioned, endonucleases are the type of nucleases involved in the cleavage of phosphodiester bonds within the nucleic acid chain (polynucleotide chain). While some of these enzymes only cut/cleave at specific sites (commonly known as restriction endonucleases), others can cut at any given site within the strand.
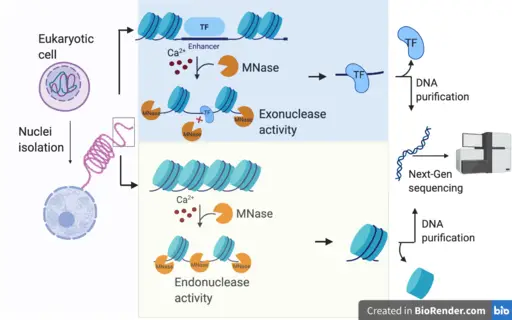
One of the other defining characteristics of endonucleases is the fact that they can cut both double-stranded and single stranded nucleic acids. This is different from exonucleases which can only cut a single strand of the molecule.
Endonucleases are often divided into two main groups that include:
· Structure-independent endonucleases
· Structure-dependent endonucleases
Structure-independent Endonucleases (sequence-independent endonucleases)
A good example of a structure/sequence-independent endonuclease is Escherichia coli endonuclease I (Endo1). As the name suggests, these endonucleases tend to randomly cleave the DNA strand (within the DNA structure). This is because they do not depend on a specific sequence or structure of the strand in order to cut.
These enzymes are primarily involved in cleaning up DNA contaminations. Because of the nature of their action, they are commonly referred to as possessive enzymes. As such, they continue to cleave DNA once the action has been initiated.
* Some examples of structure independent endonuclease that cleaves double-stranded nucleic acids (DNA) are Micrococcal nuclease and DNase 1. As well, an example of structure-independent endonuclease that acts on a single nucleic strand is mung bean nuclease.
Structure-dependent Endonucleases
Unlike structure-independent endonucleases, structure-dependent endonucleases require a specific site or damaged base in order to act. A good example of such sites is the apurinic site (apyrimidinic site) also known as AP sites. These sites occur spontaneously when a given base pair is lost and can negatively affect the cell's functions.
In such an event, the enzyme endonuclease IV jumps into action to catalyze the cleavage of the phosphodiester bonds at the site to create a 1 nucleotide gap (this cap consists of a 3' hydroxyl and 5' deoxyribose phosphate terminal). 8-oxoguanine (8-oxoG) is also an example of DNA error. This type of error in the DNA occurs when guanine is oxidized.
This error is eliminated by the enzyme Fpg. When activated, the enzyme serves to remove the damaged purines (2-6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7, 8-dihydroguanine). This action creates a 1 nucleotide gap with 5' and 3' phosphate terminals.
* Once specific sites are cleaved, the sticky ends are hybridized by DNA ligase through a process known as recombination. This results in a DNA strand known as recombinant DNA.
Some of the other types of endonucleases include:
Glycosylases - Some of the endonucleases only act as glycosylases. These endonucleases only function by removing the damaged base. They do not act on the DNA backbone. An example of these enzymes is Uracil-DNA Glycosylase.
Secondary structure endonucleases - As the name suggests, these endonucleases only act on the secondary structure of the DNA. Here, an example of the Thermostable FEN1 which identifies and cleaves a flap structure on the strand. In doing so, it leaves a 5' phosphate that can be ligated by the enzyme DNA ligase.
Restriction Endonucleases
Also known as restriction enzymes, restriction endonucleases are a type of endonucleases produced by bacteria and archaea. Like the other structure-dependent endonucleases, these enzymes are site-specific and therefore act on specific regions of the strand.
These sites are commonly known as restriction sites and consist of a short sequence of nucleotides. In most cases, the restriction sites read the same forward and backward (e.g. GTAATG) and are therefore known as palindromes/inverted repeats.
There are four main types of restriction enzymes/endonucleases which include:
Type I restriction enzymes (Type I enzymes) - The type I restriction enzyme only functions in the presence of ATP and S-adenosyl-L-methionine (C-factors). Also known as combination restriction and modification enzymes, the members of this group typically cut randomly at regions far from the recognized site (they cut about 1,000 base pairs away from the recognition site).
Type II restriction enzymes - Unlike type I enzymes, type II restriction enzymes typically cut/cleave within the recognition site or at least a short distance from the recognition site. In most cases, they function in the presence of magnesium and often produce more discrete restriction fragments - These enzymes also consist of smaller subunits when compared to type I enzymes.
Some examples of Type II enzymes include Type IIS, Type IIG, and NotI.
Type III restriction enzymes - Type III enzymes normally cleave at sites between 20 and 30 pairs away from the recognition site. Like type I enzymes, they require ATP and S-adenosyl-L-methionine to function and consist of several subunits. As compared to some of the other enzymes, type III enzymes can recognize two non-palindromic and inversely oriented sequences.
Type IV restriction enzymes - Type IV enzymes are endonucleases that recognize modified DNA (e.g. methylated DNA and Hydroxymethylated DNA). Some of the most common type IV enzymes include Mrr and McrBC systems found in E. coli.
* Being some of the most common endonucleases, restriction enzymes are often used in gene cloning and protein expression, DNA mapping, and preparing DNA libraries.
* These enzymes can cleave the double-stranded DNA, single-stranded DNA, as well as RNAs.
While these enzymes play a vital role in DNA repair, they also protect bacteria against invading viruses. This is achieved through selective cleavage of viral DNA. This action against invading viral DNA is known as restriction digestion.
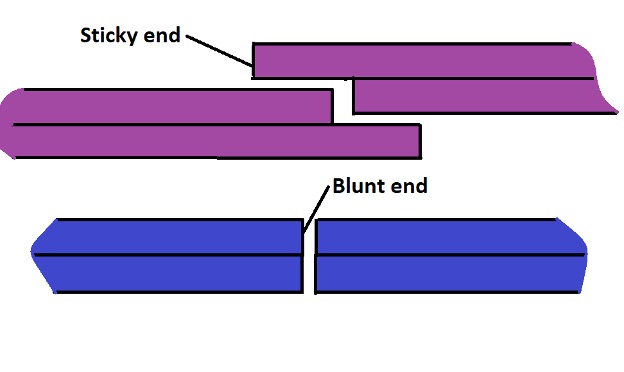
As mentioned both in DNA repair and restriction digestion, the enzymes start by recognizing the restriction site. These sites consist of a specific sequence of nucleotides - 4-5 bases. Following recognition, the site is then acted upon/cleaved. This is often uneven and involves acting on the phosphodiester bonds. Eventually, this action generates sticky or blunt ends which can then be reconnected through hybridization (through the action of DNA ligase)
* The different types of endonucleases recognize and act on specific sites.
Exonucleases: Activities and Functions
Exonucleases are nucleases that act by cutting/removing nucleotides from the ends of nucleic acid strands. There are many different types of exonucleases with different structural and biochemical characteristics. Like endonucleases, they are involved in a number of important functions including maintenance of genomic DNA integrity, DNA repair, degradation and turnover of RNA, regulating transcription, and enhancing the fidelity of DNA synthesis.
Depending on the type of exonuclease, they can remove nucleotides from the 5' to 3 direction of 3' to 5' direction (from ends of both single-stranded and double-stranded DNA strands).
Exonucleases carry out their functions by cleaving the phosphodiester bonds through a process known as hydrolysis (a chemical reaction in which water is used to break chemical bonds). Because of differences in their structure and biochemical traits, exonucleases display some differences in their activities.
For instance, the majority of exonucleases will digest DNA with nicks. Essentially, a nick is a notch/dent commonly found in double-stranded DNA molecules. These notches are often the result of lost phosphodiester bonds between neighboring nucleotides. Unlike endonucleases, this takes place at the ends of the strand. With the exception of Axo V and Exo VIII, the majority of exonucleases can digest DNA molecules with nicks.
Some of the exonucleases perform their function by removing a single base at a time (rather than a nucleotide). A good example of this is the Lambda exonuclease. When activated, the enzyme starts acting on the double-stranded DNA from the 5' end (free end) and degrades the entire strand. This activity transforms a double-stranded DNA into a single-stranded DNA.
Some of the other exonucleases that act by removing one base at a time are Exonuclease I and Exonuclease III (of E.coli). Unlike the Lambda Exonuclease, the two enzymes start acting at the 3' end of the strand.
Some of the other mechanisms of other types of exonucleases include:
Removing short oligos - Also known as oligonucleotides, oligos are short strands of DNA or RNA. Some of the exonucleases involved in the removal of these oligos include Exonuclease V, Exonuclease VII, and T5 Exonuclease. Whereas the T5 Exonuclease starts from the 5' end to the 3' end, the other two can start from any direction.
Double-stranded DNA and single-stranded DNA - Some exonucleases have been shown to act on double-stranded DNA while others can only act on a single-stranded molecule. Like Exonuclease V, the enzyme T5 exonuclease can also act on double-stranded DNA. However, Exonuclease T and Exonuclease I can only digest a single-stranded DNA and thus does not act on double-stranded molecules.
As well, Exonuclease V can digest both single-stranded and double-stranded DNA molecules.
* Compared to some of the other exonucleases, the enzyme Exonuclease V is unique in that it consists of helicase and endonuclease. It's involved in a number of functions including unwinding a double-stranded DNA molecule and DNA digestion.
Endonucleases Vs Exonucleases - Differences
Both endonucleases and exonucleases are nucleases whose primary function is to cleave nucleic acid strands (DNA/RNA). They can be found in eukaryotic and prokaryotic cells (E.g. E. coli) and achieve their function by cutting the phosphodiester bonds that hold adjacent nucleotides together.
However, they have several differences that include:
Site of action - As mentioned, one of the main difference between endonuclease and exonuclease is that endonucleases cleave the phosphodiester bonds within the nucleic acid strands while exonucleases cleave the DNA from either end (5' or 3' end).
Here, exonucleases usually need the 5' or 3' (phosphate and hydroxyl groups respectively) end in order to act on the strand. Depending on the enzymes, the activity of endonuclease enzymes can result in blunt or sticky ends. However, exonuclease activities often produce sticky ends.
Given that endonucleases cut within the nucleic acid strands, oligonucleotides are often the end products. Also, exonucleases often produce nucleotide monomers by acting on the ends of a nucleic acid strand.
Sequence specificity - One of the other differences between endonucleases and exonucleases is with regards to specificity. A good number of endonucleases are sequence-specific which means that they cut the DNA strand at given points (recognition sites). This is particularly common among a group of endonucleases known as restriction enzymes/endonucleases.
In order to perform their function, these enzymes have to identify these sites (e.g. apurinic site) on the DNA strand. If the site is not detected, then the enzyme cannot act. As well, exonucleases are random (non-specific) in their activity. Here, then, their functions depend on the type of action required at any given time (e.g. degradation of RNA).
* While exonucleases are not highly specific in their function, it's worth noting that exonucleases are often divided into two main categories depending on their mode of action.
These include:
· 5' to 3' exonucleases - These exonucleases act from the 5' to 3' direction. Essentially, this means that they recognize the phosphate group (at the 5' end) before starting their activity.
· 3' to 5' exonucleases - These exonucleases act from the 3' to the 5' direction. As such, they first identify the hydroxyl group in the nucleic strand.
Lag period - Based on a number of studies, endonucleases have been shown to undergo a lag period before producing oligonucleotides. This is not the case with exonucleases. Lag phase in endonucleases has been attributed to the fact that they are more specific than exonucleases.
Defensive functions - In bacteria, as mentioned, restriction enzymes can identify and destroy viral DNA by cleaving them. This is an important defensive strategy that prevents viral particles from proliferating (prevent viral DNA from being replicated). Unlike endonucleases, exonucleases do not have this particular function.
Return from Endonuclease Vs Exonucleases to MicroscopeMaster home
References
H Slor. 1975. Differentiation between exonucleases and endonucleases and between haplotomic and diplotomic endonucleases using 3-h-dna-coated wells of plastic depression plates as substrate.
Rongjuan Mi, Anne K. Abole and Weiguo Cao. (2010). Dissecting endonuclease and exonuclease activitiesin endonuclease V from Thermotoga maritima.
Tatsuya Nishino and Kosuke Morikawa. (2002). Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors.
Links
https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/exonuclease
https://geneticeducation.co.in/endonuclease-vs-exonuclease-10-common-differences/
Find out how to advertise on MicroscopeMaster!