What are Viruses?
Types, Structures, Replication
Definition: What are Viruses?
First described by Martinus Beijerinck, a Dutch scientist, in 1899, viruses are obligate intracellular parasites that are often described as particles. Unlike many other unicellular organisms, viruses can only replicate within living cells (e.g. in bacteria, plant and animal cells).
They also have a very simple structure and lack many of the organelles found in various prokaryotic and eukaryotic cells (e.g. mitochondria, Golgi apparatus, etc). However, each virion/viral particle consists of nucleic acid that serves to classify different types of viruses. While they cause a wide range of diseases in plants and animals, viruses can also be beneficial to man.
Some of the most common viruses include:
- Epstein–Barr virus
- Norwalk virus
- Ebola virus
- Hepatitis virus
- Rhinovirus
- Simian immunodeficiency virus
A Brief History of Virology
* Virology refers to the study of viruses.
Adolf Mayer (1876) - While investigating a disease affecting tobacco plants (which he later named tobacco mosaic disease), Adolf Mayer discovered that by rubbing sap extracted from a diseased plant to a healthy plant, the disease could be transmitted from one plant to another.
Based on his findings, he concluded that a tiny bacterium or toxin was responsible for the disease.
Dmitry Ivanovsky (1892) - While studying tobacco mosaic disease, Dmitry discovered that the disease was caused by an agent that was smaller than bacteria. Through his experiments, he noticed that this agent passed through filters that trap bacteria.
Dmitry concluded that chemical toxins released by bacteria were responsible for the disease.
Martinus Beijerinck (1898) - By following up on the work of Mayer, Martinus noticed that the agent (responsible for leaf mosaic disease) could be transmitted from one plant to another and that it passed through filters that trapped bacteria. Through further studies, Martinus also noticed that the agent grew in living cells which led him to conclude that it was actually a living organism.
Martinus called the organism "contagium vivum fluidum" (meaning a living contagious fluid)
* Since the beginning of the 20th century onwards, new studies have been able to identify different types of viruses, their biology as well as classification.
General Classification of Viruses
Generally, viruses are classified based on main factors that include:
- General morphology
- Nature of the viral genome
- Size
- Mode of replication
- Type of host
General Morphology
Helical Symmetry
Some of the viruses with a helical symmetry include mumps virus, Sendai virus, and tobacco mosaic viruses. During replication, about 2,130 identical protein subunits known as promoters assemble into a helical resulting in flexible filaments or elongated rods.
Apart from the self-assembling promoters that assemble into the helical array, the nucleic acid of these viruses has also been shown to wind into a helical form.
* The protein helix structure serves to protect the nucleic acid.
* The nucleocapsid is about 1,000nm in length and about 17nm in diameter.
Icosahedral Symmetry
Some of the viruses with an icosahedral capsid include human rhinovirus, rice yellow mottle virus, cricket paralysis virus, and porcine parvovirus. The structure consists of 20 equilateral triangular faces and 12 vertices (corners) as well as 30 edges.
Generally, the icosahedron is made up of 12 capsomers (morphological units) each consisting of 5 units located at the vertices of the capsid.
* Capsomers are the building blocks/units of the capsid in viruses.
* Some of the viruses appear spherical because they are enclosed within an envelope.
Viral Genome
Essentially, a virus may contain DNA or RNA but never both. Therefore, a given virus may be classified based on the type of genome it contains. For instance, the majority of plant viruses have been shown to contain a single strand of RNA (ssRNA).
Depending on the type of virus, then, the genome may be single-stranded RNA (ssRNA), single-stranded DNA (ssDNA), double-stranded RNA (dsRNA), or double-stranded DNA (dsDNA). These strands may appear linear, circular, or segmented which also allows for identification of given viruses.
Herpes viruses contain a linear double-stranded DNA molecule while influenza A virus carries a segmented genome (8 segments of ssRNA).
As compared to other organisms where cells may contain different genomes (e.g. the presence of DNA and mRNA in eukaryotic cells), a given group of viruses can only contain a specific type of genome thereby making viral genome of the best characteristic for classifying/identifying the virus.
- Linear ssDNA - common among members of Parvovirus family
- Circular ssDNA - common among members of Circovirus family
- Linear dsDNA - common among the majority of viruses with DNA as their genome
Size
Generally, viruses are very small in size when compared to cells (single-celled organisms or cells of higher organisms). They are about 100 to 500 times smaller than an average bacterium.
Recent studies have also identified significantly large viruses (known as giant viruses) measuring up to 700nm in diameter. Due to their different sizes, it has become possible to classify viruses based on size.
The following are different types of virus based on size:
· Mimivirus - A genus of the family Mimiviridae and consists of some of the largest viruses on earth (giant viruses). In addition to a capsid that measures about 400nm in diameter, they also possess protein filaments (of about 100nm in length) that project from the surface of the capsid. measuring about 600nm in general, they are currently the largest viruses ever identified.
· Poxvirus - Belong to the family poxviridae and measure 240nm by 300nm making them some of the largest viruses
· Rhabdovirus - Belong to the family Rhabdoviridae and measure 75nm by 180nm
· Herpesviridae - Members of this family cause infections in both human beings and animals and measure between 120 and 230nm in diameter
· Adenovirus - These are medium-sized viruses that measure between 90 and 100nm in diameter
· Parvoviruses - Members of this genus are some of the smallest viruses and measure between 23 and 28nm in diameter
* While the capsid can increase in size with the addition of non-vertext capsomers, this may cause the structure to become unstable thus increasing the risk of incorrect assembly.
Mode of Replication
The mode of replication is one of the methods used to classify viruses. Apart from the type of genome that different viruses have, studies have also shown them to differ in their modes of replication.
For the majority of viruses grouped as Class 1 viruses (this includes viruses with dsDNA), DNA is copied by using DNA polymerase of the host and replication occurs within the nucleus of the host's cell. Here, some of the viruses can influence the cells of the host to start dividing.
In other viruses, DNA polymerase of the host is not required for replication. A good example of this is Class III viruses. Unlike members of Class I, these viruses carry their own polymerase when they enter the hosts' cells. In the cell, they reside in the cytoplasm where they use this polymerase enzyme to make copies of their RNA.
Based on the mode of replication, viruses are divided into seven classes that include:
· Class I viruses - This includes members of the classes Herpesviridae and Papoviridae etc. They have a dsDNA and replicate in the nucleus of the host cells using the polymerase enzyme of the host.
· Class II viruses - Includes members of Anelloviridae and Parvoviridae etc. They contain ssDNA (circular genome) and also replicate within the nucleus of the host cell. This is achieved by a rolling circle mechanism.
· Class III viruses - This includes members of the class Rheoviridae etc. Members of this group have a dsRNA and replicate in the cytoplasm of the host cell. They have their own polymerase enzyme and therefore do not heavily rely on that of the host.
· Class IV viruses - This group includes orthomyxoviruses etc. They have a single-stranded (+) sense RNA and thus the genomic RNA has a similar sense to mRNA. They replicate in the cytoplasm of the host cells using their own enzymes (e.g. RNA-dependent RNA polymerase).
· Class V viruses - Include members of the families Orthomyxoviridae and Rhabodviridae among others. They have ssRNA and replicate in the cytoplasm. Here, they use their own polymerase to make copies of mRNA.
· Class VI viruses - Some of the viruses in this group include members of the family Metaviridae. They have a positive (+) sense ssRNA and replicate via a DNA intermediate.
Here, the RNA is first converted to DNA through reverse transcriptase which is then spliced into the genome of the host. This allows for transcription and translation of the viral genome.
· Class VII viruses - Also known as pararetroviruses, members of this group (e.g. hepatitis viruses) use reverse transcriptase during replication. Here, their genome is transcribed into mRNA by the mechanism of host cells before being converted to RNA progenome.
The progenome is then converted to DNA by reverse transcriptase. However, unlike retroviruses, members of this group do not always integrate into the genome of the host cells.
* Classification of viruses based on the mode of replication and the viral genome is known as the Baltimore classification system.
Type of Host
Classification based on the type of host they infect:
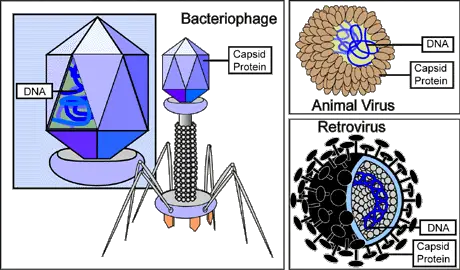
Some of the families of animal viruses include:
· Picornavirus - Small in size and include members of enteroviruses and rhinoviruses among others
· Reovirus - Cause intestinal infections in human beings and include rotaviruses
· Togaviruses - Consists of the genera alphavirus and flavivirus. They are some of the smallest enveloped viruses
· Retrovirus - They contain the enzyme reverse transcriptase and are medium-sized. They include lentivirus and have been associated with sarcomas and leukemia
· Orthomyxovirus - They measure between 80 and 90um in diameter and include the genera influenza A, B, and C
Some of the other animal viruses include:
- Paramyxovirus
- Arenavirus
- Rhabdovirus
- Parvovirus
- Papovavirus
- Adenovirus
- Poxvirus
Some of the most common plant viruses include:
- Tobacco Mosaic virus
- Tomato spotted wilt virus
- Tomato yellow leaf curl virus
- Potato virus Y
Bacteriophages
Viruses that infect bacteria:
- Enterobacteria phage T2
- Bacteriophage MS2
- Bacillus phage phi29
- Pseudomonas phage phi6
Some of the other factors that have been used to classify viruses include:
- Site of replication
- Presence or absence of an envelope
- Chemical composition
General Structure of Viruses
Viruses are not cells and are therefore often referred to as particles (virus particles/virions). Unlike cells, they have a very basic structure consisting of a few essential elements.
The simplest virus, for instance, consists of nucleic acid and a protein coat known as the capsid. Depending on the virus, the genome is either RNA or DNA. This may also be present as a single strand, double-strand, linear, circular, or segmented depending on the type of virus.
According to studies, single-stranded genomes (RNA or DNA) may have different polarities. In the event that a single strand has similar polarity to the mRNA, then they are referred to as positive (+) strands. Some of the viruses where the genome has similar polarity to the mRNA include picornaviruses and flaviviruses.
Apart from the genome, viruses also have a protein coat (capsid). The capsid covers the genome and therefore acts as a shell. It's an important component of a virion given that it protects the genome from nucleases of the host's cell.
Aside from protecting the genome, the capsid is also involved in attaching the virus to the surface of the host's cells during an infection. Proteins that make up the capsid are coded by the genome. Depending on the virus, the capsid is formed as a single or double protein shell with very few structural proteins.
* In some viruses, the nucleic acid is in direct contact with the capsid (e.g. in picornaviruses and togaviruses etc).
For more complex viruses, an enclosing structure known as the "envelope" encapsulates the particle. It resembles the cell membrane and consists of virus-specific proteins as well as lipids and carbohydrates taken from the infected cell (cell of the host).
Here, it's worth noting that the genome of viruses is limited in size and therefore codes for a few essential structural proteins. For this reason, some of the structures found in more complex viruses are taken from the cells they infect.
On the envelope, studies have shown that viral proteins may be modified to form glycoprotein by sugar groups. These glycoproteins appear as spikes (known as peplomers) on the surface or exterior of the envelope and may extend 20nm from the surface of the particle.
Here, these glycoproteins play an important role in receptor recognition thus allowing the virion to identify the appropriate host/cell. Apart from the envelope (which provides an additional layer of protection), some of the viruses have been shown to contain a tegument layer located between the capsid and envelop. This layer contains several viral protein components.
* Peplomers communicate with the matrix protein through the lipid layer.
Viral Replication
Replication of viruses results in the production of new viral genomes and structural proteins that allow the virus to multiply. Given that viruses lack many of the organelles and elements required for reproduction, they have to rely on the machinery of the host cells for replication.
Due to the fact that they are unable to replicate outside the cell of a host, viruses are described as obligate intracellular parasites.
For viruses to complete their life cycle, they first have to enter/invade the cell of a host. Depending on the virus and the type of host, this is accomplished through several means. In plants, viruses invade the cells when the cell wall is damaged (when there is a break in the cell wall).
They can also gain entry when they are injected by such sap-sucking insects as aphids. Following this initial entry into the plant cells, they are then capable of spreading to other cells through the plasmodesmata.
For animal viruses, entry into the cell starts with the virus binding to the surface of the cell. This involves the particles binding to given receptor molecules on the surface of the cell. However, these interactions may also be nonspecific, allowing certain virions to bind to any cells.
Viral receptors also actively contribute to entry through:
Initiating conformational changes of the virion
Activating the signaling pathways
Promoting endocytic internalization
Following successful binding, the capsid penetrates the membrane and the genome is released into the cytoplasm. Here, fusion proteins serve to disrupt the bilayer membrane and consequently create a pore that allows penetration.
After attachment to the cell membrane, some of the viruses are engulfed by the membrane. This occurs when viruses use signaling activities to induce changes in the cell. Here, the enveloped viruses first attaches to the receptors located on the surface of the cell membrane and stimulate the membrane to engulf the virion. This results in the formation of an endocytotic vesicle which may then fuse with lysosome.
In the vesicle, envelop proteins undergo conformational changes which causes the membrane to fuse with the endocytic membrane and consequently release the nucleocapsid into the cytoplasm. This mode of penetration is referred to as receptor-mediated endocytosis and is pH-dependent (requires acidic pH).
How Bacteriophage infect Bacteria
Bacteriophages (phage) are a group of viruses that infect bacteria. To do this, the virus first binds to specific receptors located on the host cell surface. Here, the virus/phage may bind to the proteins of lipopolysaccharides located on the outer membrane (of Gram-positive bacteria).
Once the virus is tightly bound to the cell membrane, it uses a tail-like structure located at its base to pierce through the membrane (of the bacteria) and inject its genome into the cell.
The tail-like structure in bacteriophage is complex and consists of baseplate, a non-contractible tube as well as a contractile sheath surrounding the tube. Following attachment to the cell membrane of the bacteria, the sheath contracts allowing the non-contractile tube to pierce through the membrane.
Once the virion enters the cell, the capsid is degraded by viral or host enzymes allowing the genome to be released. For viruses with an envelope, it remains in the cellular membrane of the host when the virion enters the cell.
* For some viruses, the capsid is involved in transcription and replication and is therefore not degraded.
Given that there are different types of genome for different viruses, the replication process varies from one group of viruses to another.
The following are modes of replication for DNA and RNA viruses:
DNA Viruses
dsDNA viruses (double stranded DNA viruses)
As compared to RNA viruses, most DNA viruses replicate in the nucleus of the host's cells. As such, they heavily rely on the DNA of the host as well as the host's RNA-processing machinery.
During replication, these viruses (e.g. Papovaviruses) use the DNA-dependent RNA polymerase II of the host (located in the nucleus) to transcribe viral mRNAs from the dsDNA (of the virus).
This process occurs during cell cycle making the polymerase available for viral replication. Viral RNA is then spliced and cleaved through (through cellular machinery) to produce monocistronic mRNAs that are translated in the cytoplasm.
This process is also dependent on the translation machinery of the cell. Following this translation, the dsDNA, along with essential proteins and enzymes, are packaged to form new progeny viruses.
ssDNA viruses (Single stranded DNA viruses)
These viruses (e.g. Parvoviruses) contain linear genomes and replication is dependent on the cell cycle of the host. During replication, the virus is dependent on the DNA polymerase of the host to produce ssDNA.
Using DNA-dependent RNA polymerase II of the host, they transcribe DNA to mRNA in the nucleus. Cellular splicing machinery is then used to produce mRNAs of the virus that are then translated in the cytoplasm.
RNA Viruses
dsRNA
For dsRNA viruses like Rotaviruses, the genome is segmented into several pieces/segments. During replication, the virus does not heavily rely on the enzymes of the host. As such, they carry their own RNA-dependent RNA polymerase with the replication cycle taking place in the cytoplasm (this is because they do not need elements of replication located in the nucleus).
Unlike most DNA viruses that are released from the capsid for replication to occur, dsRNA viruses like Rotaviruses retain a layer of the capsid where transcription occurs. Following transcription, mRNAs are then released into the cytoplasm where translation occurs.
The process starts with the synthesis of +ssRNA from the dsRNA segments using the RNA-dependent RNA polymerase of the virus. In the cytoplasm, some of the synthesized and capped (by capping enzymes) +ssRNAs are translated by the translation machinery of the host while the others are packed into viral capsids. As they mature, capped +ssRNAs are used as templates to produce complementary ssRNA to form dsRNA segments.
+ssRNA viruses
The RNA of these viruses (e.g. Picornaviridae and Flaviviridae etc) acts as the mRNA. During replication, the RNA-dependent RNA polymerase (of the virus) replicates the genome. Here, the genome (+ssRNA) is transcribed to produce -ssRNA that in turn serves as a template to produce genomic +ssRNA.
-ssRNA viruses
Like +ssRNA viruses, these viruses (e.g. Paramyxovirudae) produce their own RNA-dependent RNA polymerase used to transcribe the genome.
Also to read: Cell Biology, Virus Vs Bacteria
Return to Viruses under the Microscope
Return from What are Viruses? to MicroscopeMaster home
References
Dorothy H. Crawford. (2011). Viruses: A Very Short Introduction.
Fredric S. Cohen. (2016). How Viruses Invade Cells.
Jones and Bartlett Publishers. Virus Replication Cycles.
Marilyn J. Roossinck. (2016). Virus: An Illustrated Guide to 101 Incredible Microbes.
S. Modrow et al. (2013). Viruses: Definition, Structure, Classification.
Links
https://www.khanacademy.org/test-prep/mcat/cells/viruses/v/viral-replicaiton-lytic-vs-lysogenic
Find out how to advertise on MicroscopeMaster!