Recombinant DNA Technology
Steps, Applications and Gene Therapy
Overview
For all living organisms, the DNA (deoxyribonucleic acid) is the most important molecule. It carries genetic information that serves as the basis for development, growth, and reproduction among other important functions.
Information contained in this molecule is first copied, through transcription, into mRNA before being translated to produce proteins. Using this knowledge, scientists developed a new technology, Recombinant DNA technology, to combine DNA molecules from two or more organisms in order to create a new molecule of DNA that consists of new genetic combinations.
Here, scientists isolate a gene of interest from one organism and insert it into a vector thereby forming a recombinant DNA molecule. Large quantities of the gene of interest (or products of the gene) are produced when the bacteria (carrying the vector) multiplies to increase in numbers.
This is particularly important in a number of areas including medicine, in gene therapy, agriculture (to increase crop yields and increase resistance, etc), and various industries.
* Recombinant DNA - A DNA molecule that consists of two or more sequences derived from different sources.
Steps
Generally, recombinant DNA technology is a process that involves several important steps that include:
Isolation of the Gene of Interest (DNA Sequence) - Gene Therapy
As mentioned, the primary goal of recombinant DNA technology is to reproduce a gene (DNA sequence) that carries genetic combinations/information of value in medicine, agriculture, and various industries, etc. In medical science, researchers may be interested in reproducing insulin for patients with Diabetes mellitus.
Here, then, researchers have to identify the gene (INS gene) responsible for the production of insulin in human beings and isolate it in its pure form. In order to isolate the desired genes or a given sequence of the DNA molecule, it's first important to obtain DNA of the organism and purify the DNA from some of the other macromolecules like lipids and proteins, etc.
Essentially, this means that it's necessary to break the cell open in order to obtain the DNA. Depending on the type of cell, different enzymes can be used.
Whereas lysozyme is normally used to break down the cell wall of a bacterium, cellulase is used to breakdown the cell wall of plants while proteases aid in the removal of proteins that are associated with the DNA. In addition, a variety of treatment methods are also used in the process of DNA purification.
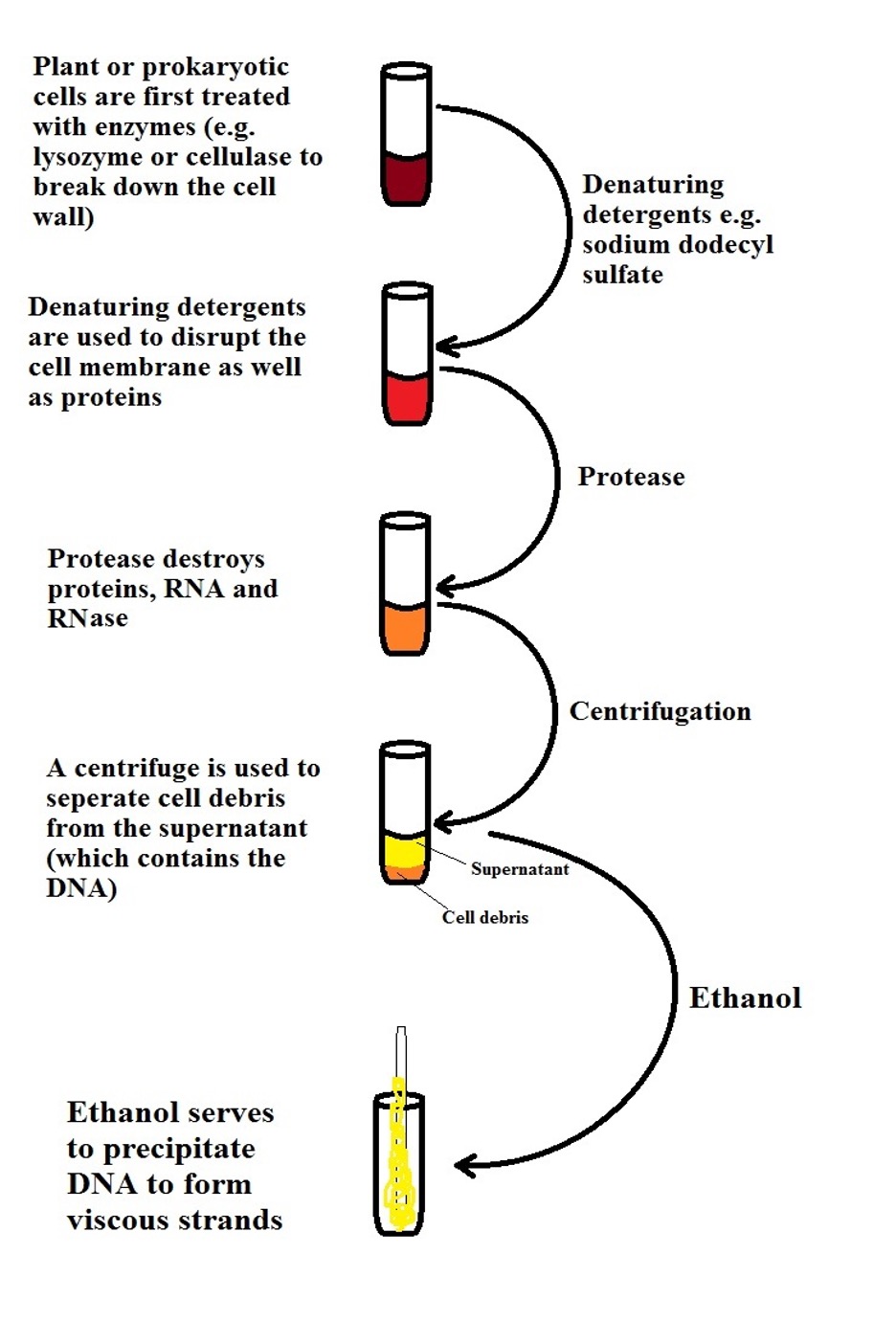
The following is a general representation of DNA isolation and purification:
* As shown in the diagram, enzymes like cellulase and lysozyme are used to break down the cell wall which surrounds the cell membrane. However, for cells that do not have a cell wall as in most eukaryotic cells, denaturing detergents are used to lyse the cells in order to free the cell contents.
Detergents like sodium dodecyl sulfate and ethyl trimethyl ammonium bromide which are surface acting agents disrupt the cell membrane allowing cell components (organelles, etc) to be released.
Once the cell membrane is destroyed (releasing contents of the cell), they are treated with the enzyme protease which acts on and destroys proteins, RNA and RNAs. Proteases have been shown to degrade enzymes and nucleases. This is particularly important as it aids in the purification of the DNA. The DNA is associated with these macromolecules under normal conditions.
Once macromolecules (proteins etc) are destroyed using various enzymes, a centrifuge is used to pellet cell debris so that they settle at the bottom of the tube while the supernatant (containing the DNA) can be extracted. Lastly, DNA is recovered through precipitation by using ethanol.
* RNAse is also used during DNA purification to degrade RNA.
Isolation of the gene of interest - Once DNA is purified; restriction enzymes are used to isolate the gene of interest. Essentially, a restriction enzyme is a type of enzyme that identifies a given sequence and cuts the DNA strand only at that particular site (the site with a specific nucleotide sequence).
Then, the enzymes act like molecular scissors given that they are able to effectively cut the DNA at specific locations to isolate the desired gene (DNA sequences e.g. INS gene).
Typically, this is achieved through the incubation of DNA (purified DNA) with a given restriction enzyme, cutting the DNA molecule at specific points, at favorable conditions that promote enzymatic activities.
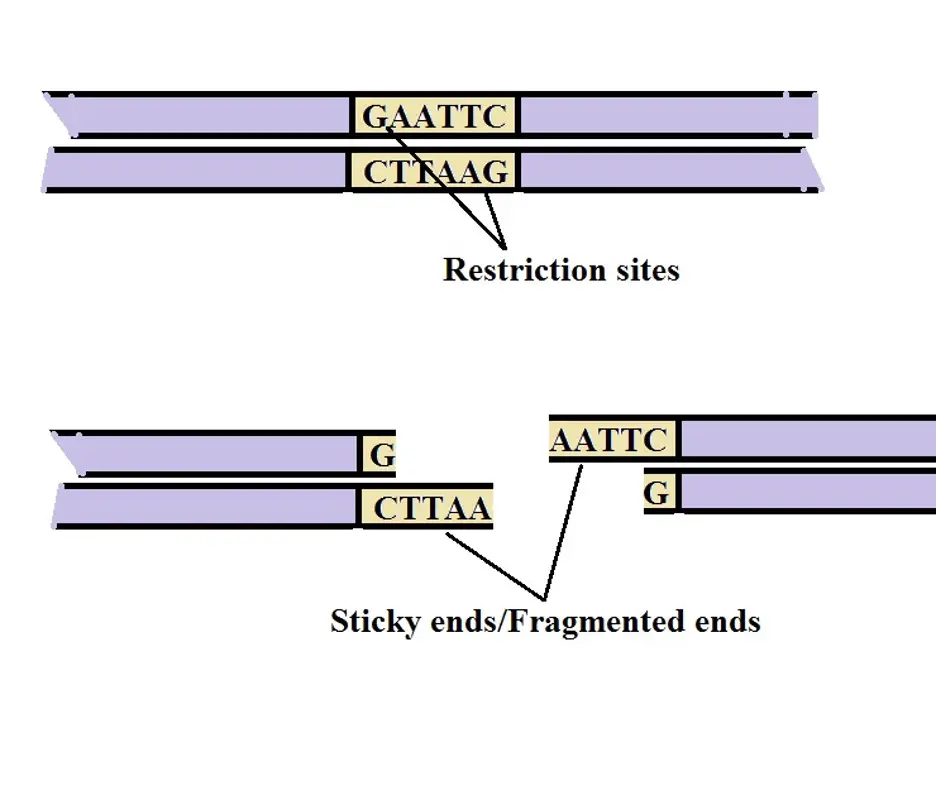
* Restriction enzyme digestion may produce fragments with staggered ends (sticky ends) where a single strand tail extends at both ends of the fragment. These ends are also known as cohesive ends and consist of base pairs that ultimately pair-up with complementary base pairs of the vector.
The following is a diagrammatic representation of the sticky ends:
* The cohesive ends allow two fragments of DNA to be joined if they were produced by the same restriction nuclease/enzyme.
* If the number of the desired gene is less than the number required for cloning, polymerase chain reaction (an amplification technique) is used to produce more copies of the gene. Using this method, it's possible to produce several thousand to millions of copies from a few gene copies.
Apart from isolating genes of interest in the laboratory, they can also be obtained from:
- Genomic library
- cDNA library
Insertion of the Isolated Gene into a Vector
The second step of recombinant DNA technology involves inserting the genes isolated in the first step into a suitable vector. Essentially, a vector is a carrier that can carry the gene of interest into a given cell where it is replicated as the cell divides.
It's worth noting that the gene of interest isolated from the DNA molecule cannot be directly introduced into a cell. This is because it may be perceived as a foreign material and destroyed. For this reason, a vector (e.g. plasmid) plays an important role in carrying the gene into the cell so that it can be replicated during normal cell division.
A vector can be described as an autonomously replicating DNA molecule. As such, it's capable of replicating on its own. A good example of a vector is a plasmid. This is a circular DNA located in the cytoplasm (commonly known as plasmid DNA).
This circular double-stranded DNA is particularly important for bacteria as it carries antibiotic-resistant genes. Apart from plasmids, bacteriophage lambda is also used as vectors.
More on Antibiotic Resistance here.
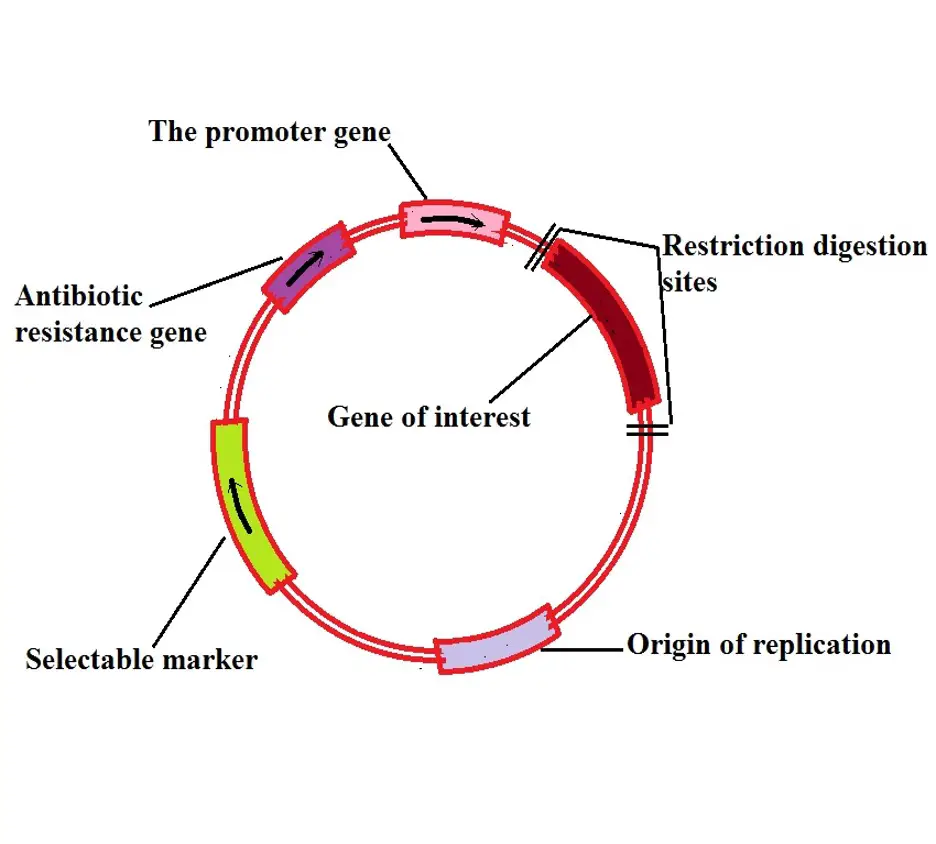
For a given vector to be used in recombinant DNA technology, it must have the following characteristics:
· Be able to self-replicate
· Have a promoter region - the region on the DNA that facilitates transcription of the target DNA
· Must have a selectable marker - important in that it allows for identification of bacteria with the gene of interest (which is carried by the plasmid) - This also makes it possible to distinguish between the bacteria and non-recombinant cells
· Have the antibiotic-resistant gene - the gene that confers antibiotic resistance to the bacteria and also makes it possible to identify the cells of interest
· Possess a cloning site - refers to the restriction endonuclease cleavage site where foreign DNA fragment is inserted
The following is a diagrammatic representation of a plasmid used in recombinant DNA technology:
In order to insert the DNA sequence/gene of interest into the vector, it's important that the ideal vector is selected and prepared. To prepare the vector (and inserts) for insertion, two important enzymes are used.
These include:
Restriction enzymes - As already mentioned, restriction enzymes serve to cleave DNA at given sites having identified specific sequences of the DNA. One of the most common restriction enzymes is EcoRI.
This enzyme is derived from E-coli and functions by recognizing the sequence 5'-GAATTC-3'. Given that there are different types of restriction enzymes, letters are used to identify their respective origins. Here, the first letter of the enzyme represents the genus in which the organism belongs while the second and third letters represent the species.
Lastly, a number is also used to indicate the order in which it was isolated from a given strain of bacteria. As mentioned, EcoR1 is one of the most common restriction enzymes. While the letter "E" represents the genus (Escherichia), the letters "co" represents the species (coli) while the last letter and number "RI" represent the strain. One of the other common enzymes is PstI which originates from the bacterium Providencia stuartii.
* Normally, bacterial restriction enzymes are produced for the purposes of protecting the cell (bacteria) against bacteriophages. When a virus inserts its DNA into a bacterium, the DNA is restricted thus preventing an infection.
* The enzyme methylase modifies restriction sites on the DNA of bacteria by adding a methyl group. This prevents restriction enzymes from cleaving their own DNA - As DNA of the phage is not methylated immediately following replication, it becomes the target of restriction enzymes.
* The type of restriction enzyme used on the plasmid will have a direct impact on some of the characteristics that the cut site confers to the bacteria. If the enzyme cuts the plasmid at the site associated with ampicillin resistance, then the gene that codes for the protein responsible for ampicillin resistance is inactivated.
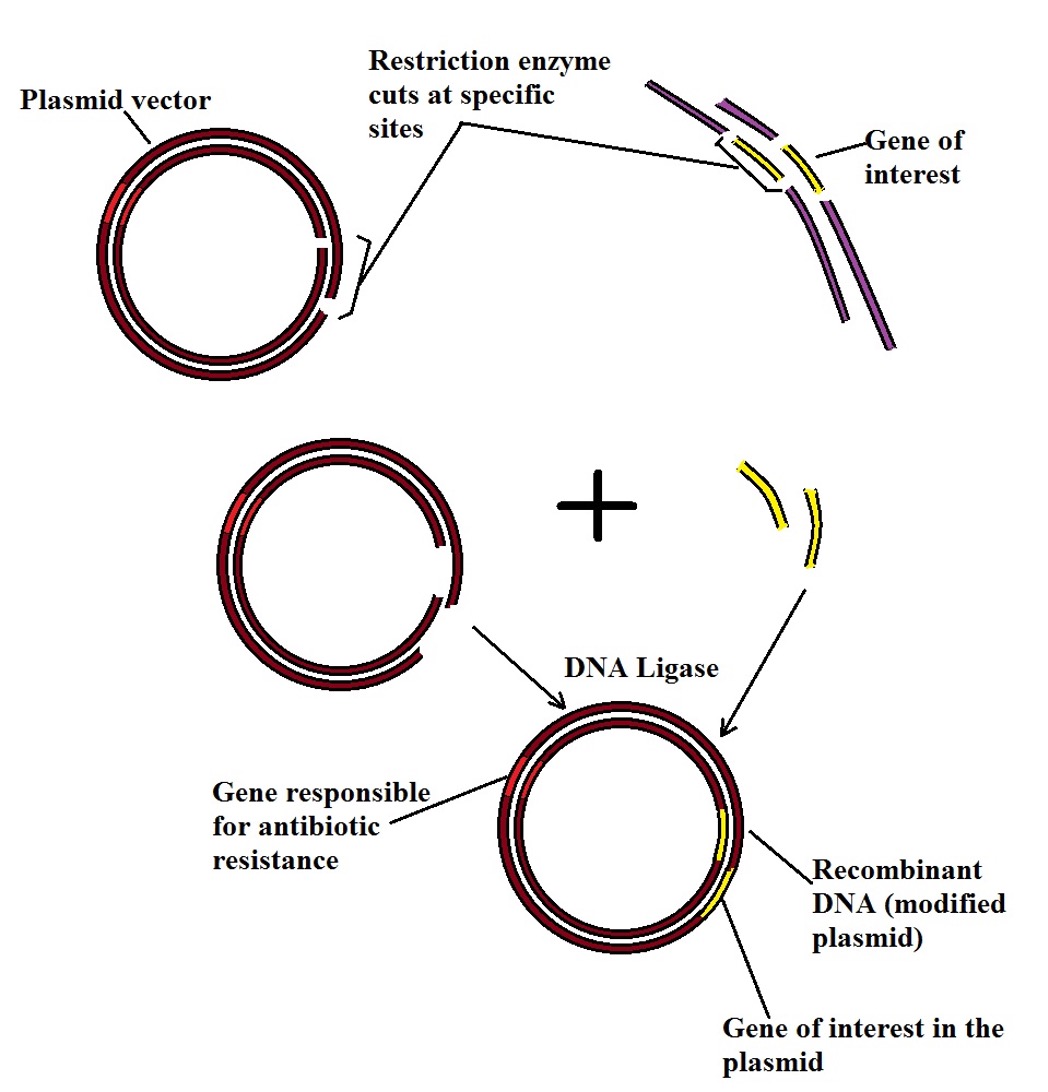
As mentioned, in order for a given vector to join with a fragment of DNA, the two must be cleaved in a manner that produces complementary ends. This becomes possible when the restriction enzyme digests both the vector and the DNA fragment to be inserted into the vector. This allows for complementary ends to be generated which ultimately join by pairing.
Although having the same enzyme digest both the vector and insert aids in the generation of complementary ends, studies have shown that the vector and insert do not necessarily have to be digested by the same enzyme for the vector and insert to join.
While the restriction enzyme is involved in cutting out the gene of interest and the vector at given sequences (the process that produces complementary ends), the two are joined by DNA ligase, a DNA-joining enzyme. However, the two can only be joined if they have matching/complementary ends.
The ligation reaction may be affected by a number of factors such as self-ligation of the vectors as well as the presence of inhibitory agents that contaminate the DNA or affect enzyme activities. For this reason, it's important to check and control these issues to ensure that ligation reactions proceed as desired.
For instance, increased concentration of vector has been shown to increase the likelihood of self-ligated vectors, so it's important to control the relative concentration of each of the two molecules (vectors and inserts). Generally, the insert vector molar ratio of 3 to 4 is said to be ideal.
Some of the other techniques used to control vector self-ligation include:
Dephosphorylation of the vector - DNA ligase requires the 5'-phosphate at the nickel site and 3'-hydroxyl group at another site for the ligation process to occur so dephosphorylation of the vector (which involves removing the 5'-phosphate group) can help reduce the chances of self-ligation
Double ingestion - This involves using two enzymes to cut the vector. This technique allows for the removal of the small fragment that exists between the cut sites thus minimizing the chances of self-ligation
Converting sticky ends to blunt ends - Given that unpaired sequences enhance the chances of ligation, trimming or filing these ends transforms them from being sticky to being blunt. This reduces the likelihood of self-ligation
Ligation reaction - The ligation reaction serves to join the insert (DNA sequence/gene of interest) and vector. Here, the enzyme joins the two DNA molecules through the formation of a phosphodiester bond using ATP energy. During this reaction, a phosphate group sticking off 5' end of one of the DNA strands is joined to the hydroxyl group that sticks off the 3' end of the other strand. In the process, a complete sugar-phosphate backbone is formed in which the insert (gene of interest) is integrated into the vector.
The following is a diagrammatic representation of restriction digestion and ligation process:
Transformation - Introduction of the Modified Vector into a host
Transformation is the third step of recombinant DNA technology and involves the introduction of the recombinant DNA (modified plasmid) into the host cell (e.g. bacterium). The primary aim of this step is to recover large amounts of the DNA molecule.
One of the most commonly used hosts is the bacterium E. coli. This is because they are inexpensive to use and have very fast growth (doubling time of about 20 minutes). Therefore, the stationary phase can be reached within a very short period of time.
Although bacterial cells like E. coli are easy to use, it's worth noting that they do not easily accept foreign genetic material. For this reason, these cells have to be treated in order to successfully introduce the new DNA.
One of the methods that was used to introduce the modified plasmid into the cell involved cooling the cells and quickly heating them up. This created little pores on the cell membrane through which the plasmids could enter the cell. Over time, however, these holes/pores would start closing up. Today, a number of new methods are being used to introduce vectors into the cells.
These include:
· Microinjection - This method involves the use of a glass needle (microcapillary pipette) to directly introduce the new DNA material into the cell. Here, a micromanipulator (for precision) is used to direct the movement of the needle.
· Electroporation - In this method, an electrical field is applied in order to increase permeability of the cell membrane. This makes it easier for the modified plasmid to be inserted into the cell.
· Ultrasound-mediated gene transfer
· Silicon Carbide fiber mediated gene transfer
· Calcium Chloride mediated transfer
* While control methods mentioned above help minimize chances of self-ligation of the vectors among other unwanted outcomes, insertion of the recombinant DNA into the host cells has been shown to result in varying outcomes.
While some of the host cells will take up the recombinant plasmid (which contains the gene of interest), some of the other host cells may take up plasmid cells that underwent self-ligation while others will contain plasmids with unwanted genes. For this reason, it's important to select transformed host cells.
Selection of Transformed Host Cells
This step of the recombinant DNA technology aims to select host cells that contain the modified recombinant vector (modified plasmid). As mentioned, some of the cells take up plasmids that underwent self-ligation while others took up plasmids with unwanted genes (some do not take in any of the plasmids). In order to achieve the desired results, it's important to only select cells that contain plasmids with the gene of interest.
Here, several methods can be used to select transformed cells. One of the most commonly used methods involves growing the cells in antibiotic (e.g. tetracycline) agar plates. In this method, media like MacConkey-Sorbitol Agar can be used to grow E. coli overnight (at 37 degrees C in neutral pH). The appropriate antibiotic is also added to the plates.
The presence of antibiotics prevents some of the cells from growing like cells that lack antibiotic resistance genes in the plasmid. However, cells with plasmids that contain antibiotic resistance genes continue growing.
Although this technique allows cells with modified genes to grow, some of the cells contain plasmids with undesired genes. Another method is needed to distinguish between the host cells that have modified plasmids (with the gene of interest) from the others (with no gene of interest).
* Gel electrophoresis is often used to separate pieces of DNA which in turn makes it possible to determine which plasmids took up the gene of interest.
Expression of the Gene introduced into the host
The last step of recombinant DNA technology is aimed at increasing the production of the desired product. Generally, recombinant DNA technology is used to increase copies of a given gene in order to increase the production of a given product. Therefore, the host cells act as factories in which the product is produced.
Under optical conditions, the host cell continues dividing which not only allows for multiplication of the recombinant DNA but also increased production through gene expression. In this step, bioreactors are used for large scale production.
Recombinant DNA Technology Applications
Today, Recombinant DNA technology is used in a number of fields including:
Agriculture - As it's now possible to introduce genes with certain desired characteristics into the DNA of another organism, recombinant DNA technology is used in agriculture to modify crops.
This has proven beneficial in a number of ways including increasing crop yield, enhancing resistance to pests, and promoting the growth and development of given plants in areas where they would otherwise not grow. By genetically modifying plants, it has become possible for researchers to end food shortages in some areas.
Medicine - In medicine, recombinant DNA technology is used for the production of various antibiotics, hormones, interferon, and vaccines, etc. For instance, using E. coli bacteria as host cells, insulin is one of the most commonly produced hormones through recombinant DNA technology.
Industry - In various industries, recombinant DNA technology is used for the purposes of producing different types of chemicals. Organic acids like citric acid are produced by microorganisms, therefore, researchers have isolated genes involved in the production of these substances for large scale production.
Gene Therapy
Gene therapy involves the introduction of given genes into the genome of an individual in order to repair mutations. Viruses are intracellular parasites and so they have been used to introduce genes into the cell for gene therapy.
In this case, the disease-causing portion of the parasite is removed in order to prevent the development of disease and introduce the therapeutic gene.
Generally, the steps highlighted above are used to prepare the parasite and use it to introduce the therapeutic gene into the cell. In the cell, the gene plays an important role in producing proteins/substances that are involved in treatment.
Although it's not widely used, recent studies are aimed at using this method to treat a wide range of diseases including diabetes, hemophilia, cancers, and cystic fibrosis among others.
Primary goals of gene therapy include:
- Replace mutated genes
- Repair mutated genes
- Promote the destruction of diseased cells by the immune system
Advantages
In recombinant DNA technology, genes with better traits are used for the purposes of overcoming various limitations. By introducing certain genes to plants, they become more resistant to given pests. Crop destruction by these pests is significantly reduced thus promoting food security. Therefore, with ongoing research recombinant presents several advantages.
Some of the other advantages associated with this technology include:
Development of vaccines and cancer treatment - Today, cancer is one of the leading causes of death. Using recombinant DNA technology, it will be possible to treat these diseases using effective genes.
On the other hand, the technology will allow for the development of better vaccines that can be used to prevent a wide range of diseases affecting the global population. In the process, it might be possible to completely eradicate some of these diseases.
Food security - As mentioned, recombinant DNA technology can be used to not only make plants resistant to pests, but also increase yields. This might be one of the most effective ways of ensuring food security across the world, and particularly in third world countries affected by food shortages and famines.
See: Endonucleases Vs Exonucleases
Return to Bacterial Conjugation
Return to DNA under the Microscope
Return from Recombinant DNA Technology to MicroscopeMaster home
References
Alberts B, Johnson A, and Lewis J, et al. (2002). Isolating, Cloning, and Sequencing DNA.
DW Ross. (1996). Tools of Recombinant DNA Technology.
Germán L. Rosano and Eduardo A. Ceccarelli. (2014). Recombinant protein expression in Escherichia coli: advances and challenges.
Keya Chaudhuri. (2013). Recombinant.
Norrgard, K. (2008) Gene therapy. Nature Education.
Sardul Singh Sandhu. (2010). Recombinant DNA Technology.
Yizhi Song et al. (2007). Ultrasound-mediated DNA transfer for bacteria.
Links
https://geneticeducation.co.in/plasmid-dna-structure-function-isolation-and-applications/
https://www.labome.com/method/DNA-Extraction-and-Purification.html
Find out how to advertise on MicroscopeMaster!