What is Antibiotic Resistance?
** Prevention
Antibiotic Resistance
Antibiotics include a group of antimicrobial substances that act against bacteria. While some of these drugs act by directly destroying bacteria, others function by inhibiting bacterial growth. In doing so, they prevent them from dividing and spreading.
While antibiotics are very effective in targeting and acting against different types of bacteria, some bacteria, for one reason or another, develop the ability to resist/withstand the actions of these substances. This is known as antibiotic resistance.
Compared to diseases/infections caused by non-resistant bacteria, those caused by antibiotic-resistant bacteria are harder to treat and are therefore associated with prolonged hospitalization and higher medical costs.
Based on the mode of action, antibiotics are classified into six major groups that include:
- Cell wall synthesis inhibitors: E.g., Penicillin and Cephalosporins
- Protein synthesis inhibitors: E.g., Macrolides, and Tetracyclines
- DNA synthesis inhibitors: E.g., Metronidazole
- RNA synthesis inhibitors: E.g., Rifampin
- Mycolic Acid synthesis inhibitors: E.g., Isoniazid
- Folic acid synthesis inhibitors: E.g., Trimethoprim
Origin of Antibiotic Resistance
Before looking at the mechanism of antibiotic resistance (how antibiotic resistance works), it's important to understand the main causes or origin of antibiotic resistance. In general, the origin of bacterial resistance can be grouped into two main categories that include natural and acquired resistance.
Natural Resistance to Antibiotics
Whereas some bacteria need to acquire certain genes in order to develop resistance to given antibiotics, there are bacteria that are naturally resistant to a given group or family of antibiotics and are therefore not affected when these antibiotics are used against them.
Here, these bacteria maybe described as having intrinsic or induced resistance to antibiotics.
Intrinsic resistance occurs when a specific group (species, genus, etc.) of bacteria exhibit traits that prevent them from being affected by given antibiotics. These traits may involve the production of enzymes that degrade antibiotics (e.g., β-lactamase), reduced permeability preventing penetration/entry of the drug into the cell, or the presence of pumps that remove toxic substances from the cell. In this case (natural resistance), bacteria have innate abilities to resist given actions of the antibiotic or a group of antibiotics.
Induced resistance, on the other hand, occurs when a given bacterium or a group of bacteria are exposed to an antibiotic. In this case, the bacterium/bacteria already possess genes that can express traits required to resist the actions of the drug. However, these genes are only expressed when the bacterium/bacteria are exposed to the antibiotic.
Following exposure to some antibiotics, studies have shown that some bacteria (e.g., E. coli) are capable of expressing multidrug efflux pumps that allow them to resist the actions of different types of antibiotics.
Unlike intrinsic resistance, where the bacterium/bacteria have already expressed traits that allow them to be resistant to given drugs, these traits are only expressed following exposure to drugs in acquired resistance.
* Inappropriate use of drugs and poor diagnostics can induce resistance to antibiotics by exposing bacteria to certain drugs.
By inappropriately using certain drugs or in the event of a misdiagnosis, drugs can activate genes responsible for resistant traits causing antibiotics to become ineffective.
However, selective pressure that causes such genes to be activated has also been shown to occur even with the appropriate use of antibiotics.
Acquired Resistance
Unlike natural resistance, acquired resistance involves the acquisition of genes that allow the bacterium/bacteria to resist the actions of given antibiotics. Here, then, bacteria do not naturally have traits or genes that allow them to be resistant.
Rather, they obtain the necessary genes from other bacteria in their surroundings which are then expressed allowing them to become resistant.
The transfer and acquisition of genes is generally known as horizontal gene transfer and may occur through any one of the following methods:
· Transformation - Transfer of DNA from a donor (exogenous DNA) to a recipient host
· Transposition - Transfer of a DNA segment from one genomic site to another
· Conjugation - A simple mating process
In nature, plasmid-mediated transfer of genes is one of the most common modes of gene transfer. Here, plasmids carrying the genes of interest are transferred and acquired by bacteria and allowing them to become resistant to given antibiotics when they are expressed. However, some bacteria can acquire these genes directly from their surroundings when other bacteria (donors) die , etc.
Once the genes enter the cell, they are integrated into the genome of the bacteria and passed down into the future generations following replication and cell division. Unlike the generation before the parent cell, the parent cell and the new generations become resistant.
* Apart from natural and acquired resistance, random mutations can cause bacteria to become resistant to various antibiotics. While most of the mutations have been shown to be of no significance or even disadvantageous to many bacteria, some of the mutations allow them to become resistant to certain antibiotics - Mutations may be the result of various factors including chemicals, ultraviolet light, or radiation, etc.
Mechanism of Antibiotic Resistance
As mentioned, there are a number of ways through which antibiotics can destroy or inhibit the growth of bacteria. However, through several mechanisms associated with the origin of resistance, some bacteria are able to overcome the effects of these drugs.
Some of the most common mechanisms of resistance include:
Enzymatic degradation of antibiotics: Inactivation of the drugs
One of the mechanisms through which some bacteria become resistant to antibiotics is by degrading or inactivating the drugs. In doing so, they make the drugs ineffective which in turn allows them to continue proliferating.
Here, one of the best examples is the ability of some bacteria to resist the effects of beta-lactam antibiotics through the production of beta-lactamase enzymes.
For many bacteria, the cell wall, which consists of peptidoglycan, is an important component that not only strengthens the cell, but also maintains the shape of the cell and also prevents the cell from bursting due to osmotic pressure.
Peptidoglycan, which is a primary component of the cell wall is made up of N-acetyl glucosamine, N-acetyl muramic acid as well as cross-linking chains of amino acid. Enzymes known as penicillin-binding proteins (transpeptidase and D-alanyl carboxypeptidase) are involved in cross-linking during the assembly of the peptidoglycan layers in the cell wall.
Beta-lactam antibiotics, which include penicillin and cephalosporins (which have a Beta-lactam ring) function by targeting the two enzymes and thus inhibiting synthesis of the cell wall. In doing so, they cause damage to the cell wall and result in the destruction of the cell because of the high internal osmotic pressure. the cell bursts because construction of the cell wall is affected.
For bacteria that are capable of producing Beta-lactamase (of those that acquire the ability to do so), they are able to deactivate the drug by breaking open the Beta-lactam ring thereby deactivating the antibiotic through a process known as hydrolysis.
* In gram-positive bacteria, the production of Beta-lactamase enzyme is induced by the presence of the antibiotic (Beta-lactam antibiotic). However, the enzyme is continually produced by Gram-negative bacteria even in cases where the bacteria has not been exposed to the antibiotic.
See also: Gram positive and Gram negative bacteria
Some of the bacteria that produce Beta-lactamase enzyme include:
- Staphylococcus aureus
- Neisseria gonorrhoeae
- Anaerobic Gram-negative bacilli
- Haemophilus influenzae
- E. coli
Alteration of drug target sites
The second mechanism through which some bacteria are able to resist the action of certain antibiotics is by altering/modifying the target sites. Different types of antibiotics function by targeting specific regions and interfering with normal biological processes. By modifying/altering these sites, the antibiotic is rendered ineffective.
Some of the most common modifications associated with antibiotic resistance include:
Altered penicillin-binding proteins - As mentioned, penicillin-binding proteins are enzymes that are involved in cross-linking of the peptidoglycan layers. In Gram-positive bacteria, the activity of these enzymes results in a thick cell wall because several layers of peptidoglycan are crosslinked.
In bacteria like Staphylococcus spp. and Streptococcus pneumoniae, alteration of these proteins contributes to resistance given that Beta-lactam antibiotics cannot bind and inhibit synthesis of the cell wall.
This may occur in cases where the bacteria acquire genes from their surroundings that produce altered proteins when they are expressed.
Despite being altered, penicillin-binding proteins are still capable of cross-linking the peptidoglycan layers. This allows for cell wall synthesis to continue while at the same time preventing Beta-lactam antibiotics from binding.
* Alteration of penicillin-binding sites increases the number of altered proteins while reducing the number of normal penicillin-binding sites. While a number of normal penicillin-binding proteins is retained, the increase in the number of altered proteins (penicillin-binding proteins) results in low-binding affinity thus reducing the effect of the beta-lactam antibiotics.
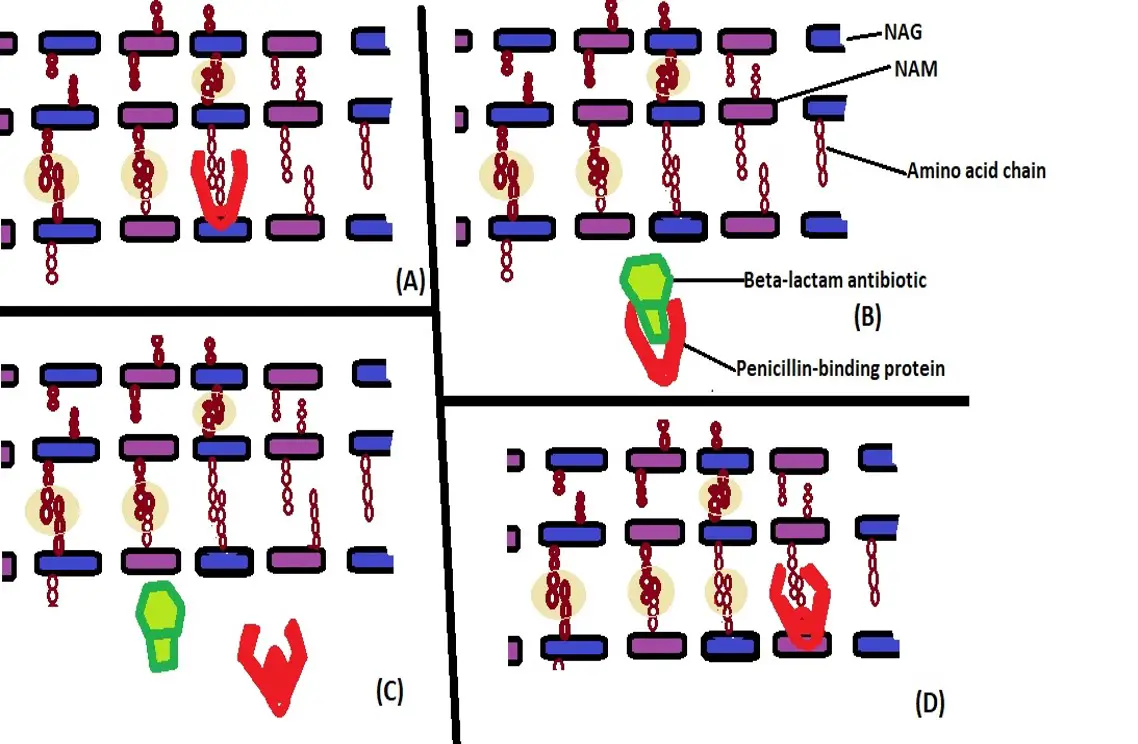
The following diagram shows how alteration of penicillin-binding proteins promote resistance to antibiotics:
(A) - Penicillin-binding protein (enzyme) linking amino acid chains between peptidoglycan layers
(B) - Beta-lactam antibiotic binds the penicillin-binding protein thus preventing cross-linking
(C) - Alteration of penicillin-binding protein prevents the Beta-lactam antibiotic from binding
(D) - Altered penicillin-binding protein continues to link amino acid chains between the peptidoglycan layers
Ribosome mutation - Ribosomes are important bacterial organelles involved in protein synthesis. This is an important process that promotes bacterial growth and division. By interfering with the activities of ribosomes, antibiotics like Macrolides are able to prevent protein synthesis and consequently bacterial growth and division.
Here, then, the ribosome (specifically the peptidyl transferase center in the large ribosome subunit) is the target site of the antibiotic action. However, as a result of a mutation at specific sites on the ribosome, antibiotics are unable to bind which renders them ineffective against the bacteria.
Mutation of the L3 ribosomal protein, which is a component of the large ribosomal subunit, is an example of ribosomal mutations that have been associated with bacterial resistance to antibiotics like tiamulin and linezolid.
These mutations have been reported in a number of bacteria including:
- E. coli
- M. tuberculosis
- Brachyspira spp.
* In addition to a mutation in the L3 ribosomal proteins, resistance has been associated with mutation of the L4 ribosomal protein as well as the methylation of ribosomal subunits.
DNA gyrase/topoisomerase IV alteration - For antibiotics like Metronidazole, interfering with DNA synthesis results in the destruction of the bacterial cell (or affects growth and division). Both DNA gyrase and topoisomerase IV are involved in DNA replication which is important during cell division.
By targeting the two enzymes (DNA gyrase and topoisomerase IV), antibiotics like Metronidazole may cause the DNA strand to break down or inhibit the replication process. In the event of a mutation of these enzymes (which causes them to chain in structure) the ability of drugs to bind is reduced which allows the bacteria to become resistant to drugs that target these enzymes.
Dihydrofolate reductase/dihydropteroate synthase mutation - DHPS and DHFR are important enzymes involved in the biosynthesis of folate in bacteria. Because of their role in this metabolic pathway, inhibiting their function (using antibiotics like Trimethoprim) can help stop bacteria growth.
As is the case with the other target sites mentioned above, antibiotics can bind to these proteins (enzymes) and prevent them from performing their function. In the event of a mutation or other changes to the structure of these proteins, antibiotics are unable to bind which allows the bacteria to develop a resistance against the drugs.
Drug efflux through efflux pumps
Found in both Gram-positive and Gram-negative bacteria, efflux pumps are important transport proteins that allow bacteria to excrete toxic substances. While some of the pumps only serve to remove specific substances from the cell, others can extrude different types of substances.
The following are different types of efflux pumps:
- ATP-binding cassette transporters (ABC)
- Major facilitator superfamily efflux pumps (MFS)
- Multidrug and toxic compound extrusion family (MATE)
- Resistance nodulation cell division family (RND)
- Small multidrug resistance family (SMR)
* Unlike some of the other mechanisms already mentioned, efflux pumps are particularly important in antibiotic resistance because they can recognize and eliminate different types of antibiotics from the cell.
For instance, whereas the MATE family efflux pump can recognize and remove aminoglycosides and fluoroquinolones from the cell, RND family efflux pumps can remove multiple types of antibiotics from the cell. In doing so, these pumps ensure that the concentration of drugs within the cell is not sufficient to cause damage.
While there are different types of efflux pumps, they can be divided into two main categories that include:
· Primary active transporters: This includes ATP binding cassette transporters
· Secondary active transporters: Includes SMR, MATE, MFS, and RND
Unlike primary active transporters, secondary active transporters use the energy produced by the chemical gradient of ions or protons.
The diagram below highlights how ATP binding cassette transporters (ABC) use ATP to extrude toxic material, including toxic antibiotics, from the cell:
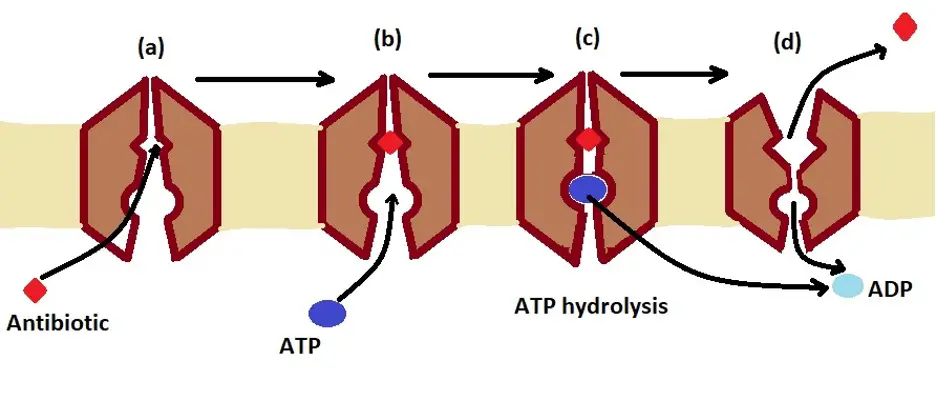
(a) - Antibiotic binds to the intermembrane protein (transporter)
(B) - ATP molecule binds to the transporter protein in the membrane and undergoes hydrolysis
(C) - ATP is hydrolyzed to produce ADP and a phosphate. This releases energy that causes the transporter protein to undergo conformational change
(D) - The protein undergoes a conformational change and releases the antibiotic outside the cell - because the ATP is hydrolyzed into ADP, it loses affinity to the protein and is released into the cell where it can undergo phosphorylation to form new ATP
* Efflux pumps allow the bacteria to remove different types of drugs that target various intracellular components and interfere with biological processes that support life.
* While efflux pumps can remove toxic materials from the intracellular environment, they can also transport material into the cell.
Limited Uptake of Drugs
While bacteria are widely divided into Gram-positive and Gram-negative bacteria, it's worth noting that some bacteria do not have a cell wall. Therefore, based on the outer envelope, bacteria can be divided into three main groups.
For bacteria with an outer membrane, Gram-negative bacteria, the high lipid content prevents hydrophilic drugs from gaining entry into the cell or limits the entry of these drugs into the cell.
As well, bacteria that lack a cell wall (e.g., Mycoplasma) are not affected by drugs that are designed to target enzymes involved in cell wall synthesis. Under certain conditions, however, given structures of the envelope can be altered which significantly limits the uptake of drugs.
Ways of Preventing Antibiotic Resistance
Antibiotic resistance can have several origins including random gene mutation and horizontal gene transfer. For these reasons, a combination of antibiotics is used to overcome resistance and treat the disease caused by bacteria. This can prove expensive due to extended hospital stay or complications resulting from the infection.
Resistance, also, can increase the risk of mortality in cases where the infection cannot be treated with available drugs. In order to prevent or at least minimize antibiotic resistance, the following preventive measures are recommended:
Finishing prescribed pills/drugs - It's important to finish taking prescribed pills as instructed by a healthcare professional. Even in a scenario where an individual feels better before completing medication, it's still important to finish the dose as instructed. This ensures that the bacterium is completely destroyed and does not develop resistance.
In cases where patients fail to finish their pills, some of the bacterial exposed to the drug may be induced to develop resistance over time. Similarly, it's important to only take antibiotics when necessary (when they are prescribed for a given infection).
Vaccination - Vaccination is important because it ensures that an individual is immunized against given infections. Here, vaccinations support the immune system and prepare it to fight off infections (e.g., whooping cough , etc.).
Proper use of antibiotics - In addition to drug abuse by human beings, there have been reports where some farmers give antibiotics to animals which contributes to resistance. It's therefore important to ensure that these drugs are only used to treat specific infections.
See more on Bacteria:
Bacteriology as a field of study
Bacterial Transformation, Conjugation
Bacteria - Size, Shape and Arrangement - Eubacteria
How do Bacteria cause Disease?
How do Antibiotics kill Bacteria?
Return from How Antibiotic Resistance happens and how to Prevent it to MicroscopeMaster home
References
Atin Sharma, Vivek Kumar Gupta, and Ranjana Pathania. (2017). Efflux pump inhibitors for bacterial pathogens: From bench to bedside.
Jun Lin, Kunihiko Nishino et al. (2015). Mechanisms of antibiotic resistance.
Matthew Metz and David M. Shlaes. (2014). Eight More Ways To Deal with Antibiotic Resistance.
Paula Blanco et al. (2016). Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants.
Wanda C Reygaert. (2018). An overview of the antimicrobial resistance mechanisms of bacteria.
Links
https://www.nfid.org/antibiotic-resistance/how-to-prevent-antibiotic-resistance/
https://www.niaid.nih.gov/research/antimicrobial-resistance-causes
Find out how to advertise on MicroscopeMaster!